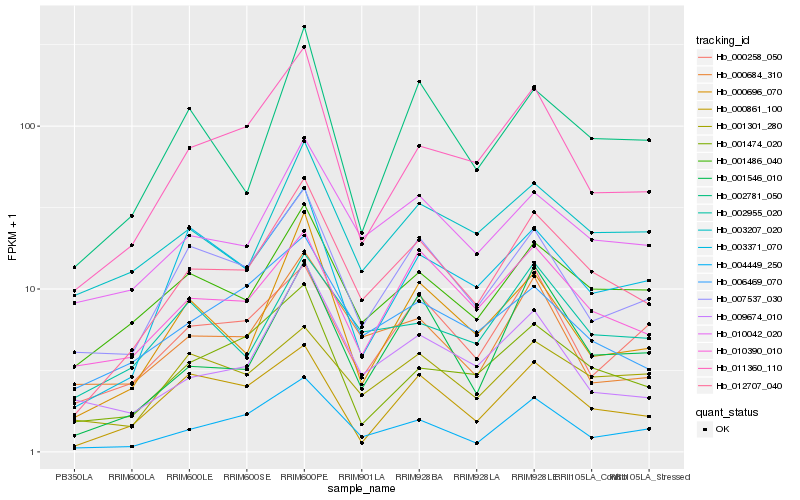
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012707_040 |
0.0 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 29 [Jatropha curcas] |
| 2 |
Hb_010042_020 |
0.1142768148 |
- |
- |
UDP-n-acteylglucosamine pyrophosphorylase, putative [Ricinus communis] |
| 3 |
Hb_001486_040 |
0.1145807219 |
- |
- |
PREDICTED: uncharacterized protein LOC105632624 isoform X1 [Jatropha curcas] |
| 4 |
Hb_007537_030 |
0.1240399207 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_004449_250 |
0.1246390698 |
- |
- |
PREDICTED: protein EI24 homolog [Jatropha curcas] |
| 6 |
Hb_011360_110 |
0.1248354015 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1 [Vitis vinifera] |
| 7 |
Hb_010390_010 |
0.1275169122 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 8 |
Hb_006469_070 |
0.1354864641 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g22040 [Jatropha curcas] |
| 9 |
Hb_000684_310 |
0.1387752185 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 10 |
Hb_000696_070 |
0.139555478 |
- |
- |
PREDICTED: aspartic proteinase-like protein 1 [Jatropha curcas] |
| 11 |
Hb_002955_020 |
0.1406761887 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_001546_010 |
0.1409647536 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003207_020 |
0.14154899 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 14 |
Hb_009674_010 |
0.1430324171 |
- |
- |
AAEL007687-PA [Aedes aegypti] |
| 15 |
Hb_003371_070 |
0.1430529504 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_001474_020 |
0.1446689073 |
- |
- |
PREDICTED: uncharacterized protein LOC105650917 [Jatropha curcas] |
| 17 |
Hb_002781_050 |
0.1450907276 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |
| 18 |
Hb_001301_280 |
0.1459960439 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000258_050 |
0.1470162216 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription repressor VAL2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000861_100 |
0.1496847155 |
- |
- |
PREDICTED: probable fucosyltransferase 8 isoform X1 [Jatropha curcas] |