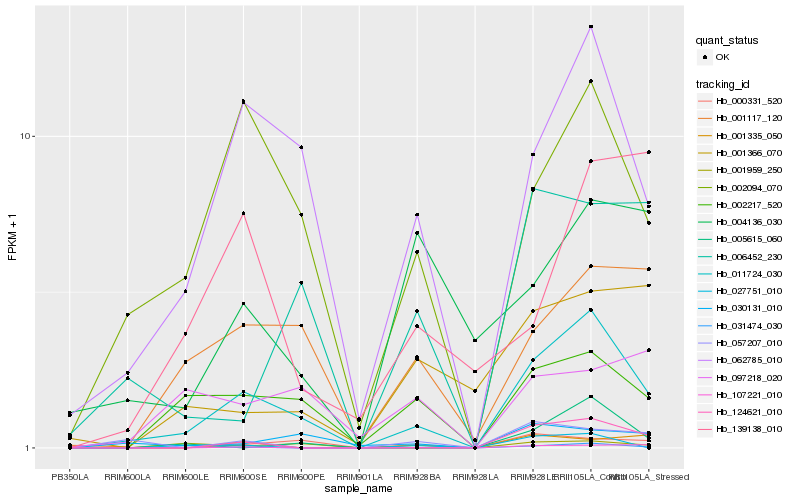
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011724_030 |
0.0 |
- |
- |
Organic cation transporter, putative [Ricinus communis] |
| 2 |
Hb_062785_010 |
0.1936999579 |
- |
- |
Organic cation transporter, putative [Ricinus communis] |
| 3 |
Hb_031474_030 |
0.2045355282 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 4 |
Hb_002094_070 |
0.2248112622 |
- |
- |
Organic cation transporter, putative [Ricinus communis] |
| 5 |
Hb_002217_520 |
0.2459027286 |
- |
- |
PREDICTED: probable protein phosphatase 2C 42 isoform X3 [Populus euphratica] |
| 6 |
Hb_001117_120 |
0.2718313478 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 7 |
Hb_000331_520 |
0.2878145289 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 8 |
Hb_124621_010 |
0.2895673549 |
- |
- |
hypothetical protein L484_019698 [Morus notabilis] |
| 9 |
Hb_030131_010 |
0.2924208971 |
- |
- |
PREDICTED: probable acyl-activating enzyme 1, peroxisomal, partial [Jatropha curcas] |
| 10 |
Hb_006452_230 |
0.3070725302 |
- |
- |
PREDICTED: uncharacterized protein LOC105635044 [Jatropha curcas] |
| 11 |
Hb_107221_010 |
0.3141348192 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 12 |
Hb_005615_060 |
0.3221946546 |
- |
- |
PREDICTED: uncharacterized protein LOC105636752 [Jatropha curcas] |
| 13 |
Hb_001959_250 |
0.3246024825 |
- |
- |
hypothetical protein CARUB_v10015435mg [Capsella rubella] |
| 14 |
Hb_097218_020 |
0.3291621308 |
- |
- |
PREDICTED: replication protein A 70 kDa DNA-binding subunit E [Jatropha curcas] |
| 15 |
Hb_057207_010 |
0.3310188152 |
- |
- |
hypothetical protein F383_17010 [Gossypium arboreum] |
| 16 |
Hb_001366_070 |
0.3330023424 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 4-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_139138_010 |
0.3330536196 |
- |
- |
PREDICTED: uncharacterized protein LOC105650977 [Jatropha curcas] |
| 18 |
Hb_004136_030 |
0.3420583151 |
- |
- |
- |
| 19 |
Hb_027751_010 |
0.3429460667 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 20 |
Hb_001335_050 |
0.3433258023 |
- |
- |
- |