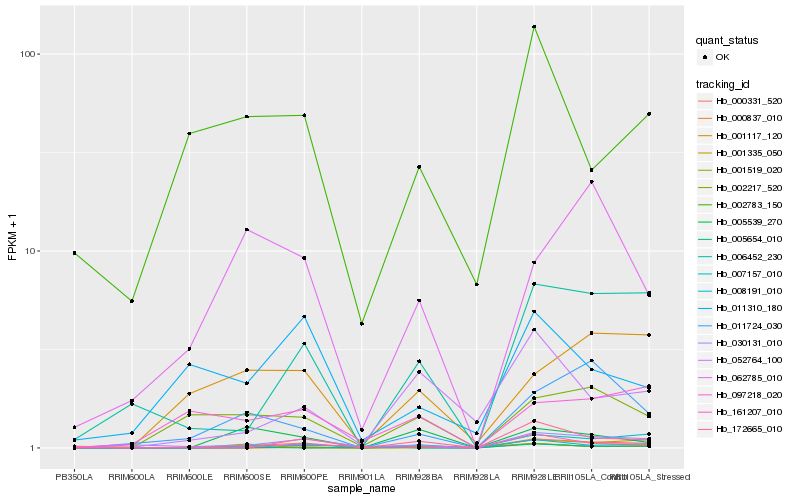
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000331_520 |
0.0 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 2 |
Hb_172665_010 |
0.2339258799 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 3 |
Hb_030131_010 |
0.23754026 |
- |
- |
PREDICTED: probable acyl-activating enzyme 1, peroxisomal, partial [Jatropha curcas] |
| 4 |
Hb_006452_230 |
0.2387323049 |
- |
- |
PREDICTED: uncharacterized protein LOC105635044 [Jatropha curcas] |
| 5 |
Hb_007157_010 |
0.2574484577 |
- |
- |
- |
| 6 |
Hb_161207_010 |
0.2827323867 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 7 |
Hb_011724_030 |
0.2878145289 |
- |
- |
Organic cation transporter, putative [Ricinus communis] |
| 8 |
Hb_002217_520 |
0.2996564897 |
- |
- |
PREDICTED: probable protein phosphatase 2C 42 isoform X3 [Populus euphratica] |
| 9 |
Hb_001117_120 |
0.3041989353 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 10 |
Hb_052764_100 |
0.3044439183 |
- |
- |
PREDICTED: beta-galactosidase 8-like [Jatropha curcas] |
| 11 |
Hb_008191_010 |
0.3063985641 |
- |
- |
- |
| 12 |
Hb_005539_270 |
0.3072152307 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 13 |
Hb_001519_020 |
0.3161794168 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 14 |
Hb_062785_010 |
0.3228167235 |
- |
- |
Organic cation transporter, putative [Ricinus communis] |
| 15 |
Hb_097218_020 |
0.3266285795 |
- |
- |
PREDICTED: replication protein A 70 kDa DNA-binding subunit E [Jatropha curcas] |
| 16 |
Hb_001335_050 |
0.3281608826 |
- |
- |
- |
| 17 |
Hb_011310_180 |
0.3360305733 |
- |
- |
PREDICTED: phospholipase D alpha 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000837_010 |
0.3366726864 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_005654_010 |
0.3374564209 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 20 |
Hb_002783_150 |
0.3391246533 |
- |
- |
cyclophilin [Hevea brasiliensis] |