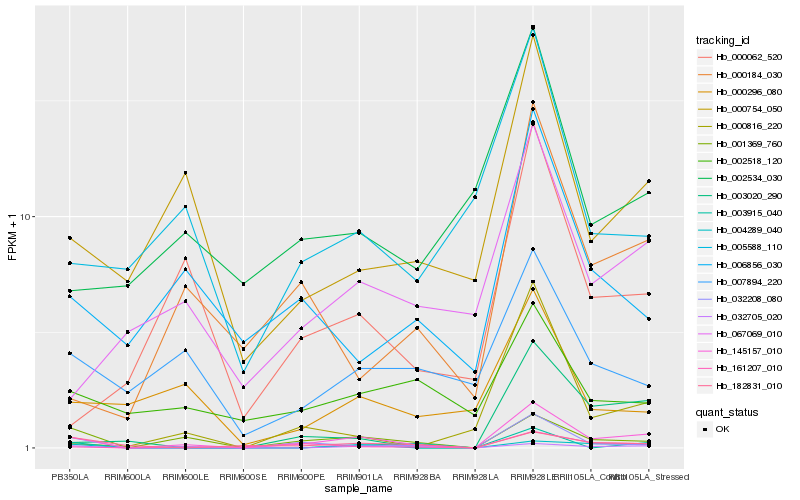
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_145157_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 2 |
Hb_032208_080 |
0.216163575 |
- |
- |
PREDICTED: phytoene synthase 2, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_001369_760 |
0.2266751987 |
- |
- |
hypothetical protein PRUPE_ppa019417mg [Prunus persica] |
| 4 |
Hb_161207_010 |
0.2601825922 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 5 |
Hb_032705_020 |
0.2756401853 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |
| 6 |
Hb_000754_050 |
0.2815772502 |
- |
- |
PREDICTED: thylakoid lumenal 19 kDa protein, chloroplastic [Jatropha curcas] |
| 7 |
Hb_004289_040 |
0.2975294996 |
- |
- |
PREDICTED: uncharacterized protein LOC104234330 [Nicotiana sylvestris] |
| 8 |
Hb_003020_290 |
0.2982925138 |
- |
- |
hypothetical protein CICLE_v10029104mg [Citrus clementina] |
| 9 |
Hb_000184_030 |
0.2996956147 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 10 |
Hb_067069_010 |
0.3050445384 |
- |
- |
hypothetical protein JCGZ_11524 [Jatropha curcas] |
| 11 |
Hb_000062_520 |
0.3053239611 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_006856_030 |
0.3076048737 |
- |
- |
Uridine kinase-like protein 4 [Glycine soja] |
| 13 |
Hb_007894_220 |
0.3113025081 |
- |
- |
PREDICTED: probable folate-biopterin transporter 4 [Jatropha curcas] |
| 14 |
Hb_003915_040 |
0.3128678775 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At2g30600 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000296_080 |
0.3133721491 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g35130 [Jatropha curcas] |
| 16 |
Hb_182831_010 |
0.3134772382 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 17 |
Hb_000816_220 |
0.3154284553 |
- |
- |
PREDICTED: CBL-interacting protein kinase 18-like [Jatropha curcas] |
| 18 |
Hb_005588_110 |
0.3176373235 |
- |
- |
hypothetical protein POPTR_0002s12860g [Populus trichocarpa] |
| 19 |
Hb_002534_030 |
0.3226829864 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 20 |
Hb_002518_120 |
0.3230442691 |
- |
- |
hypothetical protein RCOM_0731250 [Ricinus communis] |