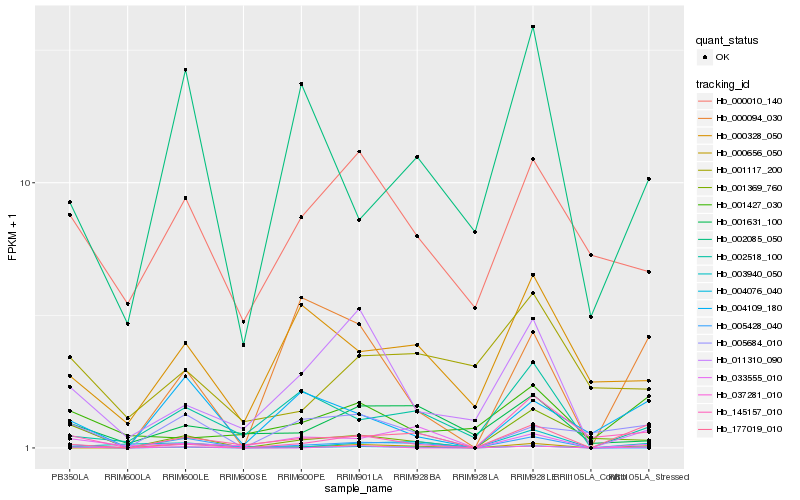
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004076_040 |
0.0 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 2 |
Hb_177019_010 |
0.2553481194 |
- |
- |
Uncharacterized protein TCM_004012 [Theobroma cacao] |
| 3 |
Hb_001369_760 |
0.2678951758 |
- |
- |
hypothetical protein PRUPE_ppa019417mg [Prunus persica] |
| 4 |
Hb_005684_010 |
0.2983964339 |
- |
- |
PREDICTED: uncharacterized protein LOC104216799 [Nicotiana sylvestris] |
| 5 |
Hb_037281_010 |
0.3042951469 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 6 |
Hb_005428_040 |
0.3080938177 |
- |
- |
- |
| 7 |
Hb_011310_090 |
0.318555576 |
- |
- |
PREDICTED: putative ATPase N2B isoform X3 [Vitis vinifera] |
| 8 |
Hb_001631_100 |
0.3264311037 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_033555_010 |
0.3280679442 |
- |
- |
- |
| 10 |
Hb_003940_050 |
0.3287396728 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 11 |
Hb_000094_030 |
0.3351985072 |
- |
- |
- |
| 12 |
Hb_001427_030 |
0.3369364023 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000010_140 |
0.3373388632 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 11, chloroplastic isoform X2 [Jatropha curcas] |
| 14 |
Hb_145157_010 |
0.3390718048 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 15 |
Hb_004109_180 |
0.3398562027 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g14410 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000328_050 |
0.3451003759 |
- |
- |
PREDICTED: translation initiation factor IF-3, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002518_100 |
0.345663947 |
- |
- |
- |
| 18 |
Hb_000656_050 |
0.345998186 |
- |
- |
PREDICTED: 3-oxoacyl-[acyl-carrier-protein] synthase II, chloroplastic-like [Vitis vinifera] |
| 19 |
Hb_001117_200 |
0.3528854685 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002085_050 |
0.3538704669 |
- |
- |
PREDICTED: beta-galactosidase 8 [Jatropha curcas] |