| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
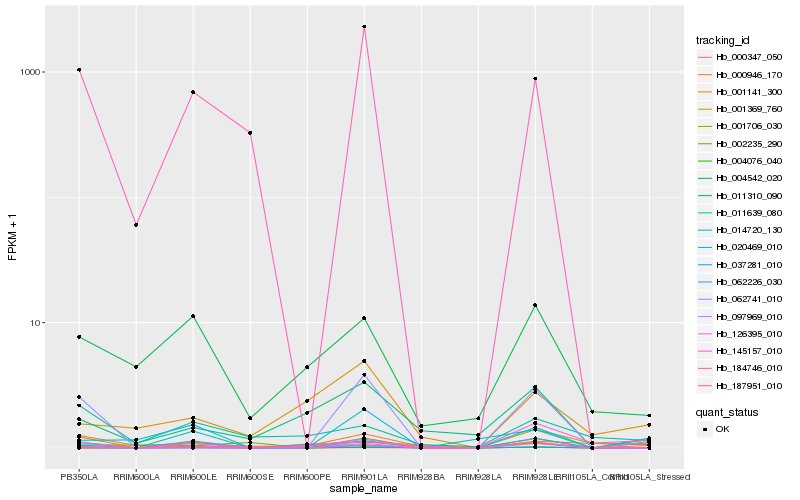
Hb_037281_010 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 2 |
Hb_187951_010 |
0.2838259873 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 3 |
Hb_004076_040 |
0.3042951469 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 4 |
Hb_184746_010 |
0.3462087056 |
- |
- |
HSP20-like chaperones superfamily protein [Theobroma cacao] |
| 5 |
Hb_020469_010 |
0.3467202292 |
- |
- |
- |
| 6 |
Hb_002235_290 |
0.3627859581 |
- |
- |
PREDICTED: casparian strip membrane protein 5 [Jatropha curcas] |
| 7 |
Hb_014720_130 |
0.3670113986 |
- |
- |
PREDICTED: uncharacterized protein LOC105640368 [Jatropha curcas] |
| 8 |
Hb_097969_010 |
0.375184274 |
- |
- |
F-box/LRR-repeat protein, putative [Ricinus communis] |
| 9 |
Hb_004542_020 |
0.378113877 |
- |
- |
hypothetical chloroplast RF1 [Hevea brasiliensis] |
| 10 |
Hb_000347_050 |
0.3844926183 |
- |
- |
PREDICTED: ABC transporter B family member 3-like [Jatropha curcas] |
| 11 |
Hb_000946_170 |
0.3950838159 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase-like [Jatropha curcas] |
| 12 |
Hb_001369_760 |
0.3971328043 |
- |
- |
hypothetical protein PRUPE_ppa019417mg [Prunus persica] |
| 13 |
Hb_001706_030 |
0.4043111697 |
- |
- |
PREDICTED: uncharacterized protein LOC104880364 [Vitis vinifera] |
| 14 |
Hb_062741_010 |
0.407341692 |
- |
- |
- |
| 15 |
Hb_011310_090 |
0.4102384442 |
- |
- |
PREDICTED: putative ATPase N2B isoform X3 [Vitis vinifera] |
| 16 |
Hb_001141_300 |
0.4125633329 |
- |
- |
- |
| 17 |
Hb_126395_010 |
0.415325635 |
- |
- |
hypothetical protein VITISV_043980 [Vitis vinifera] |
| 18 |
Hb_145157_010 |
0.4217467965 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 19 |
Hb_062226_030 |
0.4257119764 |
- |
- |
- |
| 20 |
Hb_011639_080 |
0.4290455331 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: receptor-like protein kinase FERONIA [Populus euphratica] |