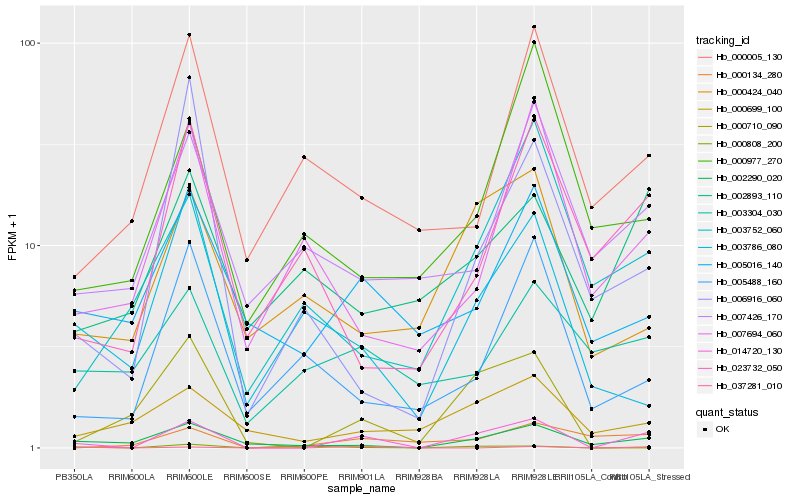
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014720_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640368 [Jatropha curcas] |
| 2 |
Hb_002290_020 |
0.3194324108 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000808_200 |
0.347464342 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase CKI1-like [Jatropha curcas] |
| 4 |
Hb_005016_140 |
0.3594013106 |
- |
- |
PREDICTED: thylakoid lumenal 15.0 kDa protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_037281_010 |
0.3670113986 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 6 |
Hb_000699_100 |
0.3680820591 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g01510, mitochondrial-like [Jatropha curcas] |
| 7 |
Hb_000134_280 |
0.3686513359 |
- |
- |
PREDICTED: uncharacterized protein LOC105628677 [Jatropha curcas] |
| 8 |
Hb_000424_040 |
0.3687087377 |
- |
- |
PREDICTED: uncharacterized protein LOC105638080 [Jatropha curcas] |
| 9 |
Hb_023732_050 |
0.3705317052 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 10 |
Hb_000710_090 |
0.3725327343 |
- |
- |
PREDICTED: uncharacterized protein LOC104583249 [Brachypodium distachyon] |
| 11 |
Hb_006916_060 |
0.373337702 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 12 |
Hb_003752_060 |
0.3736211141 |
- |
- |
PREDICTED: epoxide hydrolase 3 [Jatropha curcas] |
| 13 |
Hb_007426_170 |
0.3755201419 |
transcription factor |
TF Family: TCP |
- |
| 14 |
Hb_005488_160 |
0.376182185 |
- |
- |
PREDICTED: maltose excess protein 1, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_007694_060 |
0.3776389391 |
- |
- |
PREDICTED: 50S ribosomal protein L35, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002893_110 |
0.3778966683 |
transcription factor |
TF Family: DBP |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |
| 17 |
Hb_003786_080 |
0.379757718 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 18 |
Hb_000005_130 |
0.3835521094 |
- |
- |
hypothetical protein POPTR_0004s22660g [Populus trichocarpa] |
| 19 |
Hb_003304_030 |
0.3859522395 |
- |
- |
PREDICTED: uroporphyrinogen-III synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 20 |
Hb_000977_270 |
0.3862958926 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 1B, chloroplastic [Jatropha curcas] |