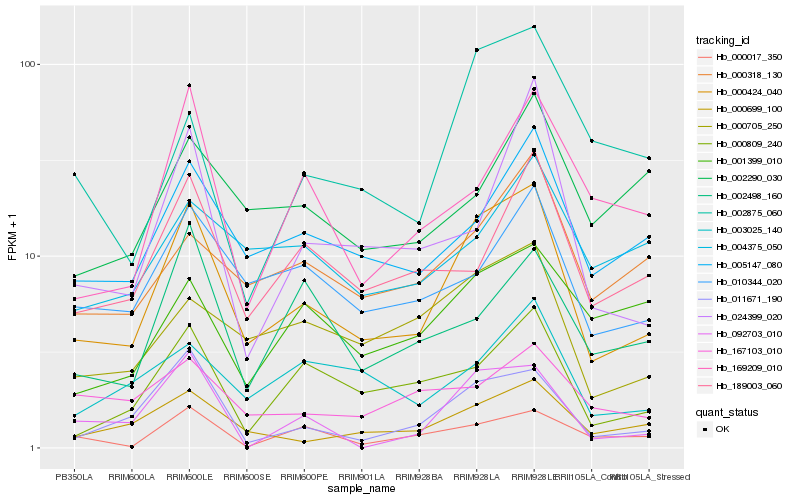
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000424_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638080 [Jatropha curcas] |
| 2 |
Hb_011671_190 |
0.1613468541 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 3 |
Hb_000699_100 |
0.1615644897 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g01510, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_005147_080 |
0.1677594125 |
- |
- |
PREDICTED: prolycopene isomerase, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_000705_250 |
0.1779501909 |
- |
- |
hypothetical protein CICLE_v10017607mg [Citrus clementina] |
| 6 |
Hb_010344_020 |
0.1797414205 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g79490, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000809_240 |
0.1816568127 |
- |
- |
hypothetical protein POPTR_0007s11280g [Populus trichocarpa] |
| 8 |
Hb_189003_060 |
0.187039636 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_000318_130 |
0.1941876637 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_003025_140 |
0.1967272094 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_169209_010 |
0.1968460312 |
- |
- |
PREDICTED: porphobilinogen deaminase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002875_060 |
0.2016786888 |
- |
- |
hypothetical protein L484_023461 [Morus notabilis] |
| 13 |
Hb_002498_160 |
0.2066388701 |
- |
- |
PREDICTED: uncharacterized protein LOC105649315 [Jatropha curcas] |
| 14 |
Hb_024399_020 |
0.2069181789 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 15 |
Hb_000017_350 |
0.2071671 |
- |
- |
PREDICTED: uncharacterized protein LOC105645799 isoform X2 [Jatropha curcas] |
| 16 |
Hb_092703_010 |
0.2098875084 |
- |
- |
PREDICTED: putative kinase-like protein TMKL1 [Jatropha curcas] |
| 17 |
Hb_001399_010 |
0.2113343792 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_004375_050 |
0.2125054937 |
- |
- |
PREDICTED: putative GTP diphosphokinase RSH1, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_167103_010 |
0.2129260988 |
- |
- |
PREDICTED: transcriptional activator DEMETER isoform X1 [Jatropha curcas] |
| 20 |
Hb_002290_030 |
0.2130052594 |
- |
- |
PREDICTED: uncharacterized protein LOC105635877 isoform X1 [Jatropha curcas] |