| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
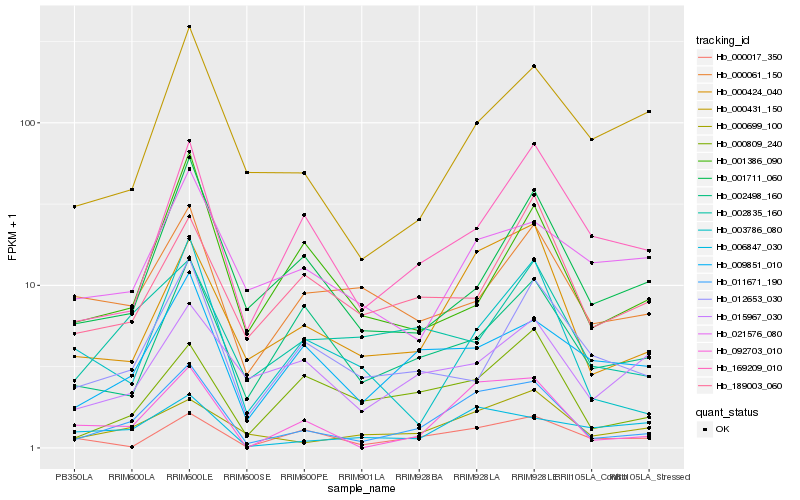
Hb_011671_190 |
0.0 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 2 |
Hb_000424_040 |
0.1613468541 |
- |
- |
PREDICTED: uncharacterized protein LOC105638080 [Jatropha curcas] |
| 3 |
Hb_092703_010 |
0.1614468587 |
- |
- |
PREDICTED: putative kinase-like protein TMKL1 [Jatropha curcas] |
| 4 |
Hb_000699_100 |
0.2027609636 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g01510, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_009851_010 |
0.214930612 |
- |
- |
PREDICTED: beta-amylase 3, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_169209_010 |
0.2195450888 |
- |
- |
PREDICTED: porphobilinogen deaminase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000809_240 |
0.2235731673 |
- |
- |
hypothetical protein POPTR_0007s11280g [Populus trichocarpa] |
| 8 |
Hb_003786_080 |
0.2283177855 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 9 |
Hb_001711_060 |
0.2293948124 |
- |
- |
PREDICTED: ferredoxin isoform X1 [Jatropha curcas] |
| 10 |
Hb_000431_150 |
0.2305215164 |
- |
- |
ribosomal protein L27, putative [Ricinus communis] |
| 11 |
Hb_002498_160 |
0.2340583949 |
- |
- |
PREDICTED: uncharacterized protein LOC105649315 [Jatropha curcas] |
| 12 |
Hb_002835_160 |
0.2373564924 |
- |
- |
Kinase superfamily protein [Theobroma cacao] |
| 13 |
Hb_006847_030 |
0.2384693888 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_012653_030 |
0.2427506592 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 4, chloroplastic isoform X2 [Jatropha curcas] |
| 15 |
Hb_000017_350 |
0.2470902513 |
- |
- |
PREDICTED: uncharacterized protein LOC105645799 isoform X2 [Jatropha curcas] |
| 16 |
Hb_015967_030 |
0.2471966777 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription repressor VAL1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_189003_060 |
0.2486150689 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_021576_080 |
0.2487976175 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY1 [Jatropha curcas] |
| 19 |
Hb_000061_150 |
0.2494485081 |
- |
- |
PREDICTED: ribulose-phosphate 3-epimerase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001386_090 |
0.2497391607 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |