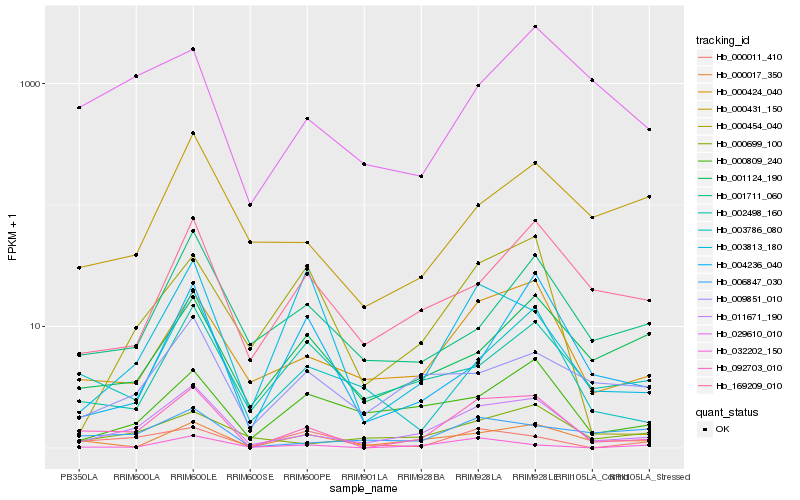
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_092703_010 |
0.0 |
- |
- |
PREDICTED: putative kinase-like protein TMKL1 [Jatropha curcas] |
| 2 |
Hb_011671_190 |
0.1614468587 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 3 |
Hb_000424_040 |
0.2098875084 |
- |
- |
PREDICTED: uncharacterized protein LOC105638080 [Jatropha curcas] |
| 4 |
Hb_003786_080 |
0.24382378 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 5 |
Hb_000017_350 |
0.245861908 |
- |
- |
PREDICTED: uncharacterized protein LOC105645799 isoform X2 [Jatropha curcas] |
| 6 |
Hb_169209_010 |
0.2774909549 |
- |
- |
PREDICTED: porphobilinogen deaminase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_032202_150 |
0.2782772979 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR14 [Jatropha curcas] |
| 8 |
Hb_002498_160 |
0.282693041 |
- |
- |
PREDICTED: uncharacterized protein LOC105649315 [Jatropha curcas] |
| 9 |
Hb_006847_030 |
0.283226972 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_000011_410 |
0.2870680443 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_029610_010 |
0.2900305519 |
- |
- |
caffeic acid 3-O-methyltransferase [Hevea brasiliensis] |
| 12 |
Hb_003813_180 |
0.2901046103 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 13 |
Hb_000454_040 |
0.2923529022 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 14 |
Hb_000809_240 |
0.2923534285 |
- |
- |
hypothetical protein POPTR_0007s11280g [Populus trichocarpa] |
| 15 |
Hb_001711_060 |
0.2933108604 |
- |
- |
PREDICTED: ferredoxin isoform X1 [Jatropha curcas] |
| 16 |
Hb_009851_010 |
0.2934518507 |
- |
- |
PREDICTED: beta-amylase 3, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_004236_040 |
0.2937432849 |
- |
- |
biotin carboxyl carrier protein [Vernicia fordii] |
| 18 |
Hb_000699_100 |
0.2958734536 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g01510, mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_001124_190 |
0.2962479251 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000431_150 |
0.2981843787 |
- |
- |
ribosomal protein L27, putative [Ricinus communis] |