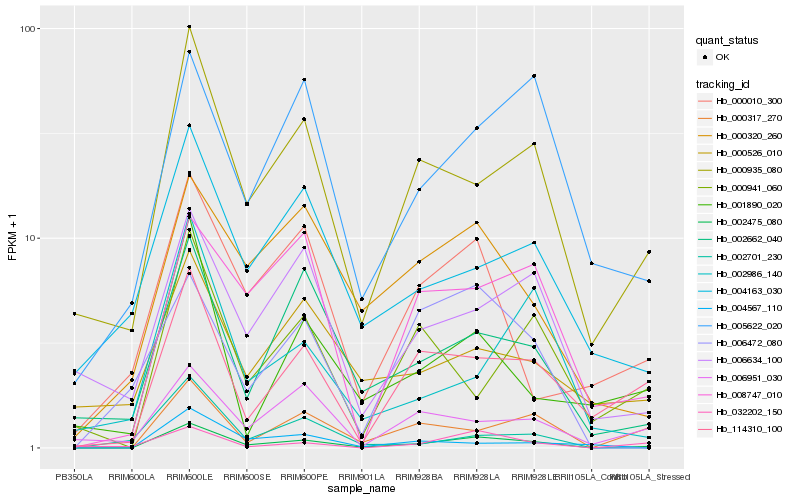
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002475_080 |
0.0 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein MALE MEIOCYTE DEATH 1 [Jatropha curcas] |
| 2 |
Hb_000935_080 |
0.196555928 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_114310_100 |
0.2071522461 |
- |
- |
glucose-1-phosphate adenylyltransferase, putative [Ricinus communis] |
| 4 |
Hb_000317_270 |
0.2166896583 |
- |
- |
Uncharacterized protein TCM_001673 [Theobroma cacao] |
| 5 |
Hb_002662_040 |
0.2251110605 |
- |
- |
PREDICTED: uncharacterized protein LOC105630915 [Jatropha curcas] |
| 6 |
Hb_008747_010 |
0.2412572798 |
- |
- |
PREDICTED: protein argonaute 2-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000320_260 |
0.2421170436 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_032202_150 |
0.2438774554 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR14 [Jatropha curcas] |
| 9 |
Hb_006634_100 |
0.2448310231 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_005622_020 |
0.2465148433 |
- |
- |
PREDICTED: DCC family protein At1g52590, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001890_020 |
0.249307449 |
- |
- |
hypothetical protein CICLE_v10025198mg [Citrus clementina] |
| 12 |
Hb_000010_300 |
0.2518489912 |
transcription factor |
TF Family: TAZ |
protein binding protein, putative [Ricinus communis] |
| 13 |
Hb_004163_030 |
0.2579182219 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A10 [Jatropha curcas] |
| 14 |
Hb_006951_030 |
0.2587530496 |
- |
- |
PREDICTED: protein MCM10 homolog [Jatropha curcas] |
| 15 |
Hb_000941_060 |
0.2595569555 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0016s05890g [Populus trichocarpa] |
| 16 |
Hb_006472_080 |
0.2601987227 |
- |
- |
- |
| 17 |
Hb_002701_230 |
0.2666454982 |
- |
- |
PREDICTED: uncharacterized protein LOC105646833 [Jatropha curcas] |
| 18 |
Hb_000526_010 |
0.2667086297 |
- |
- |
PREDICTED: uroporphyrinogen-III C-methyltransferase [Jatropha curcas] |
| 19 |
Hb_004567_110 |
0.2675251821 |
- |
- |
hypothetical protein POPTR_0002s24610g [Populus trichocarpa] |
| 20 |
Hb_002986_140 |
0.2693722514 |
- |
- |
PREDICTED: uncharacterized protein LOC105639684 [Jatropha curcas] |