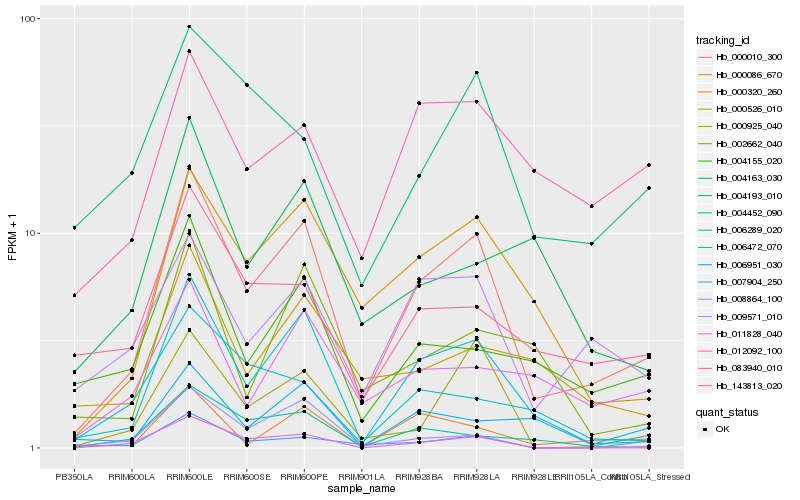
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000010_300 |
0.0 |
transcription factor |
TF Family: TAZ |
protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_000320_260 |
0.1814961092 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_009571_010 |
0.1928367462 |
- |
- |
early-responsive to dehydration family protein [Populus trichocarpa] |
| 4 |
Hb_004452_090 |
0.1951323789 |
- |
- |
hypothetical protein JCGZ_12231 [Jatropha curcas] |
| 5 |
Hb_000086_670 |
0.2086532405 |
- |
- |
PREDICTED: probable DNA helicase MCM9 [Jatropha curcas] |
| 6 |
Hb_006472_070 |
0.2105607239 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 7 |
Hb_006289_020 |
0.21140115 |
- |
- |
PREDICTED: shugoshin-1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004155_020 |
0.2128181233 |
transcription factor |
TF Family: MYB |
myb, putative [Ricinus communis] |
| 9 |
Hb_143813_020 |
0.212974624 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0006s10950g [Populus trichocarpa] |
| 10 |
Hb_007904_250 |
0.2134068646 |
- |
- |
PREDICTED: vignain-like [Jatropha curcas] |
| 11 |
Hb_000526_010 |
0.2165117908 |
- |
- |
PREDICTED: uroporphyrinogen-III C-methyltransferase [Jatropha curcas] |
| 12 |
Hb_011828_040 |
0.2182614176 |
- |
- |
PREDICTED: uncharacterized protein LOC105642173 isoform X2 [Jatropha curcas] |
| 13 |
Hb_004193_010 |
0.2226543901 |
- |
- |
PREDICTED: uncharacterized protein LOC105650669 [Jatropha curcas] |
| 14 |
Hb_006951_030 |
0.2233331611 |
- |
- |
PREDICTED: protein MCM10 homolog [Jatropha curcas] |
| 15 |
Hb_083940_010 |
0.2243665418 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 16 |
Hb_012092_100 |
0.2253487311 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002662_040 |
0.2314574442 |
- |
- |
PREDICTED: uncharacterized protein LOC105630915 [Jatropha curcas] |
| 18 |
Hb_000925_040 |
0.2320394317 |
- |
- |
kinase, putative [Ricinus communis] |
| 19 |
Hb_004163_030 |
0.2345236625 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A10 [Jatropha curcas] |
| 20 |
Hb_008864_100 |
0.2364891376 |
- |
- |
PREDICTED: uncharacterized protein LOC105631832 isoform X1 [Jatropha curcas] |