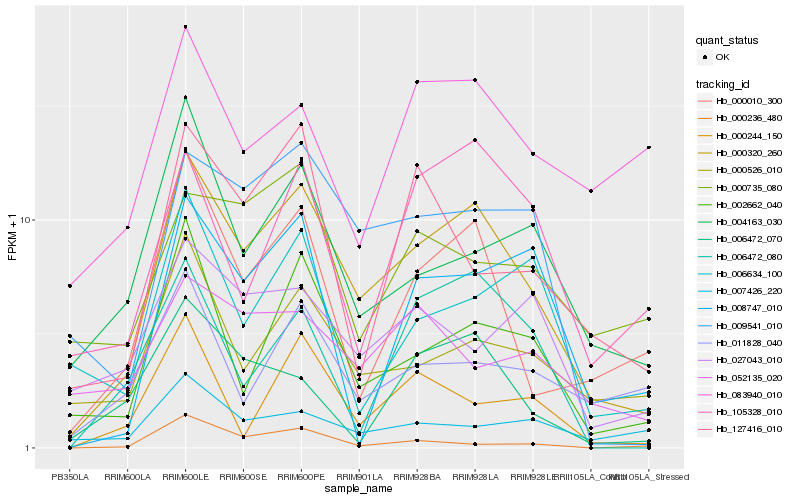
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000320_260 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_000010_300 |
0.1814961092 |
transcription factor |
TF Family: TAZ |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_009541_010 |
0.1882913391 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 6 [Jatropha curcas] |
| 4 |
Hb_002662_040 |
0.1921742329 |
- |
- |
PREDICTED: uncharacterized protein LOC105630915 [Jatropha curcas] |
| 5 |
Hb_000526_010 |
0.2002184294 |
- |
- |
PREDICTED: uroporphyrinogen-III C-methyltransferase [Jatropha curcas] |
| 6 |
Hb_004163_030 |
0.2050205985 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A10 [Jatropha curcas] |
| 7 |
Hb_105328_010 |
0.2199315081 |
- |
- |
PREDICTED: low-temperature-induced cysteine proteinase-like [Jatropha curcas] |
| 8 |
Hb_011828_040 |
0.2239124006 |
- |
- |
PREDICTED: uncharacterized protein LOC105642173 isoform X2 [Jatropha curcas] |
| 9 |
Hb_127416_010 |
0.2243137483 |
- |
- |
LysM domain GPI-anchored protein 2 precursor, putative [Ricinus communis] |
| 10 |
Hb_006472_070 |
0.2258903864 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 11 |
Hb_008747_010 |
0.2261045067 |
- |
- |
PREDICTED: protein argonaute 2-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_027043_010 |
0.228790553 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase GDPD4 [Jatropha curcas] |
| 13 |
Hb_006634_100 |
0.2318828852 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000244_150 |
0.2325606189 |
- |
- |
PREDICTED: chromatin assembly factor 1 subunit FAS2 [Jatropha curcas] |
| 15 |
Hb_006472_080 |
0.2326231581 |
- |
- |
- |
| 16 |
Hb_007426_220 |
0.2332882715 |
- |
- |
Endoplasmic oxidoreductin-1 precursor, putative [Ricinus communis] |
| 17 |
Hb_052135_020 |
0.2340475295 |
- |
- |
PREDICTED: MAP kinase kinase MKK1/SSP32-like [Jatropha curcas] |
| 18 |
Hb_000735_080 |
0.2353263196 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 6 [Jatropha curcas] |
| 19 |
Hb_000236_480 |
0.2375383634 |
- |
- |
PREDICTED: cytochrome P450 CYP72A219-like [Jatropha curcas] |
| 20 |
Hb_083940_010 |
0.2397476545 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |