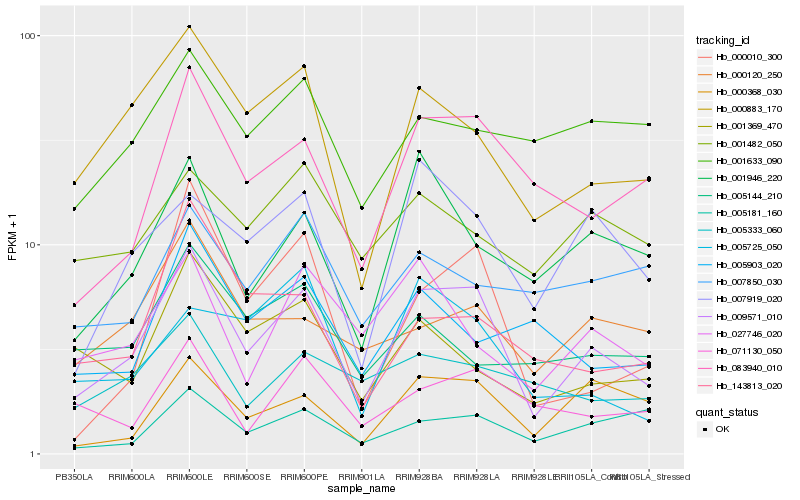
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009571_010 |
0.0 |
- |
- |
early-responsive to dehydration family protein [Populus trichocarpa] |
| 2 |
Hb_000883_170 |
0.1609672289 |
- |
- |
senescence-associated family protein [Populus trichocarpa] |
| 3 |
Hb_083940_010 |
0.163551828 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 4 |
Hb_007919_020 |
0.1838051569 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 5 |
Hb_001946_220 |
0.1864791543 |
- |
- |
PREDICTED: signal peptidase complex catalytic subunit SEC11A-like [Jatropha curcas] |
| 6 |
Hb_000368_030 |
0.1895094174 |
- |
- |
PREDICTED: cytokinin dehydrogenase 5 [Jatropha curcas] |
| 7 |
Hb_000010_300 |
0.1928367462 |
transcription factor |
TF Family: TAZ |
protein binding protein, putative [Ricinus communis] |
| 8 |
Hb_027746_020 |
0.1969894444 |
- |
- |
PREDICTED: replication protein A 70 kDa DNA-binding subunit B [Jatropha curcas] |
| 9 |
Hb_071130_050 |
0.1977835566 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 10 |
Hb_000120_250 |
0.198551866 |
- |
- |
PREDICTED: uncharacterized protein LOC105630152 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001369_470 |
0.2001496767 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKS1 [Jatropha curcas] |
| 12 |
Hb_005333_060 |
0.2018553944 |
- |
- |
PREDICTED: uncharacterized protein LOC105630688 [Jatropha curcas] |
| 13 |
Hb_143813_020 |
0.2042320179 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0006s10950g [Populus trichocarpa] |
| 14 |
Hb_005181_160 |
0.2051501447 |
- |
- |
PREDICTED: uncharacterized protein LOC105643546 [Jatropha curcas] |
| 15 |
Hb_005903_020 |
0.2087935049 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 16 |
Hb_005725_050 |
0.2102223152 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Jatropha curcas] |
| 17 |
Hb_001482_050 |
0.210354954 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 18 |
Hb_001633_090 |
0.2107057122 |
- |
- |
PREDICTED: mitochondrial phosphate carrier protein 3, mitochondrial [Jatropha curcas] |
| 19 |
Hb_005144_210 |
0.2132868157 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_007850_030 |
0.2157666151 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |