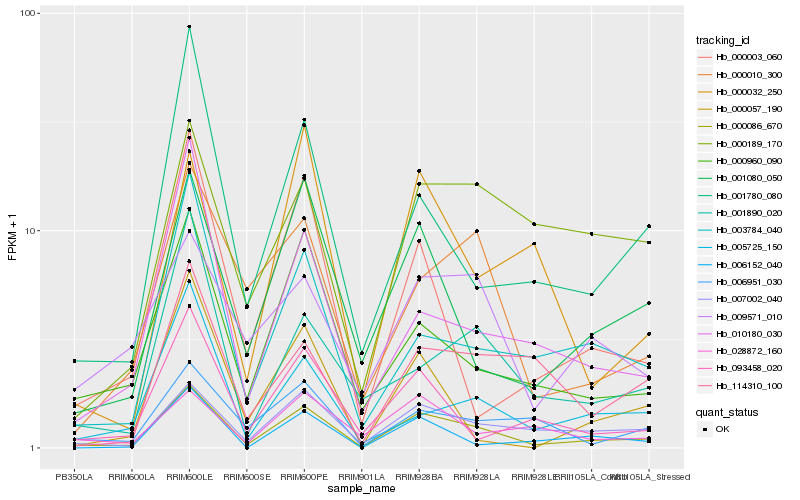
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000086_670 |
0.0 |
- |
- |
PREDICTED: probable DNA helicase MCM9 [Jatropha curcas] |
| 2 |
Hb_007002_040 |
0.1535141049 |
transcription factor |
TF Family: PHD |
DNA replication regulator dpb11, putative [Ricinus communis] |
| 3 |
Hb_001080_050 |
0.1877847682 |
- |
- |
PREDICTED: uncharacterized protein LOC105635235 [Jatropha curcas] |
| 4 |
Hb_000960_090 |
0.1924933228 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003784_040 |
0.2027360798 |
- |
- |
Afadin- and alpha-actinin-binding protein, putative [Ricinus communis] |
| 6 |
Hb_000010_300 |
0.2086532405 |
transcription factor |
TF Family: TAZ |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_001780_080 |
0.2115022471 |
- |
- |
PREDICTED: tubulin beta-2 chain [Jatropha curcas] |
| 8 |
Hb_006951_030 |
0.2129849442 |
- |
- |
PREDICTED: protein MCM10 homolog [Jatropha curcas] |
| 9 |
Hb_010180_030 |
0.2139398367 |
- |
- |
PREDICTED: WEB family protein At2g38370-like [Jatropha curcas] |
| 10 |
Hb_093458_020 |
0.2139944857 |
desease resistance |
Gene Name: AAA |
PREDICTED: chromosome transmission fidelity protein 18 homolog [Jatropha curcas] |
| 11 |
Hb_000003_060 |
0.2184347032 |
transcription factor |
TF Family: HMG |
DNA-binding protein MNB1B, putative [Ricinus communis] |
| 12 |
Hb_006152_040 |
0.2187548553 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105645062 isoform X2 [Jatropha curcas] |
| 13 |
Hb_005725_150 |
0.2188556141 |
- |
- |
PREDICTED: kinesin-related protein 11 isoform X1 [Jatropha curcas] |
| 14 |
Hb_114310_100 |
0.2204486546 |
- |
- |
glucose-1-phosphate adenylyltransferase, putative [Ricinus communis] |
| 15 |
Hb_028872_160 |
0.2252540369 |
- |
- |
PREDICTED: ATP-dependent DNA helicase Q-like 5 [Jatropha curcas] |
| 16 |
Hb_000189_170 |
0.2253631968 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 17 |
Hb_000057_190 |
0.2268837894 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001890_020 |
0.2283509964 |
- |
- |
hypothetical protein CICLE_v10025198mg [Citrus clementina] |
| 19 |
Hb_000032_250 |
0.2284442274 |
- |
- |
PREDICTED: endoglucanase 6 [Jatropha curcas] |
| 20 |
Hb_009571_010 |
0.2290921232 |
- |
- |
early-responsive to dehydration family protein [Populus trichocarpa] |