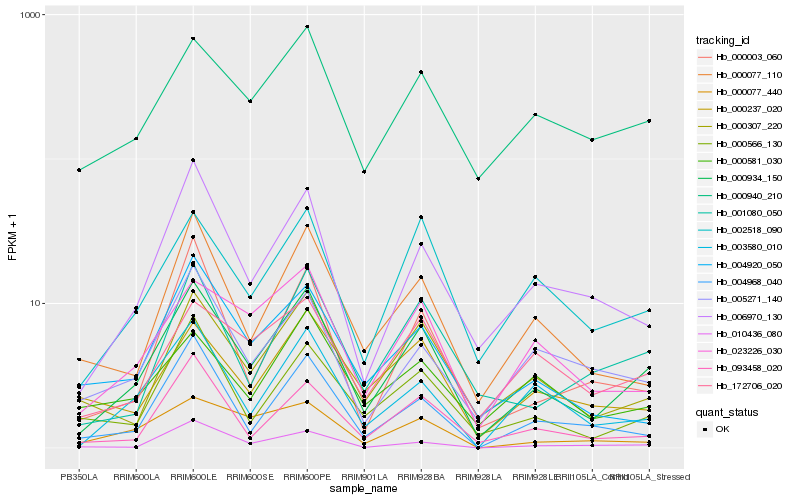
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000934_150 |
0.0 |
- |
- |
hypothetical protein POPTR_0005s21000g [Populus trichocarpa] |
| 2 |
Hb_000307_220 |
0.1500688741 |
- |
- |
oxidoreductase family protein [Populus trichocarpa] |
| 3 |
Hb_002518_090 |
0.1515851187 |
- |
- |
PREDICTED: tubulin beta-2 chain [Jatropha curcas] |
| 4 |
Hb_006970_130 |
0.1545796755 |
- |
- |
PREDICTED: histone H3.3-like [Colobus angolensis palliatus] |
| 5 |
Hb_003580_010 |
0.1568485384 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 6 |
Hb_023226_030 |
0.1611712098 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-14-like [Jatropha curcas] |
| 7 |
Hb_001080_050 |
0.1615964245 |
- |
- |
PREDICTED: uncharacterized protein LOC105635235 [Jatropha curcas] |
| 8 |
Hb_000566_130 |
0.1629667668 |
- |
- |
PREDICTED: probable DNA primase large subunit isoform X2 [Jatropha curcas] |
| 9 |
Hb_172706_020 |
0.1651262695 |
- |
- |
PREDICTED: protein ABIL2 [Jatropha curcas] |
| 10 |
Hb_000940_210 |
0.1661246794 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 11 |
Hb_000077_110 |
0.1679899436 |
- |
- |
PREDICTED: uncharacterized protein LOC105646935 [Jatropha curcas] |
| 12 |
Hb_093458_020 |
0.1690382434 |
desease resistance |
Gene Name: AAA |
PREDICTED: chromosome transmission fidelity protein 18 homolog [Jatropha curcas] |
| 13 |
Hb_005271_140 |
0.1764480078 |
transcription factor |
TF Family: LIM |
Pollen-specific protein SF3 [Morus notabilis] |
| 14 |
Hb_000077_440 |
0.1803998266 |
- |
- |
PREDICTED: ELMO domain-containing protein A isoform X1 [Jatropha curcas] |
| 15 |
Hb_004920_050 |
0.1806104964 |
- |
- |
DNA replication licensing factor MCM7, putative [Ricinus communis] |
| 16 |
Hb_000003_060 |
0.1818675191 |
transcription factor |
TF Family: HMG |
DNA-binding protein MNB1B, putative [Ricinus communis] |
| 17 |
Hb_004968_040 |
0.1822813637 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 5 [Jatropha curcas] |
| 18 |
Hb_000581_030 |
0.1857692098 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000237_020 |
0.186175142 |
- |
- |
PREDICTED: DNA replication licensing factor MCM4 [Jatropha curcas] |
| 20 |
Hb_010436_080 |
0.1889563631 |
- |
- |
PREDICTED: DNA replication licensing factor MCM5 [Jatropha curcas] |