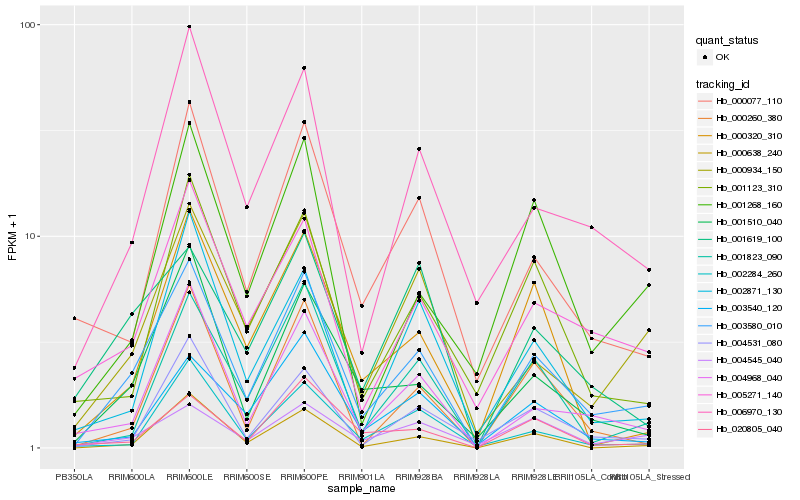
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003580_010 |
0.0 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 2 |
Hb_004968_040 |
0.1495331133 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 5 [Jatropha curcas] |
| 3 |
Hb_006970_130 |
0.1557757351 |
- |
- |
PREDICTED: histone H3.3-like [Colobus angolensis palliatus] |
| 4 |
Hb_000934_150 |
0.1568485384 |
- |
- |
hypothetical protein POPTR_0005s21000g [Populus trichocarpa] |
| 5 |
Hb_001510_040 |
0.158048591 |
- |
- |
PREDICTED: 125 kDa kinesin-related protein [Jatropha curcas] |
| 6 |
Hb_001823_090 |
0.1625915303 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005271_140 |
0.1652734962 |
transcription factor |
TF Family: LIM |
Pollen-specific protein SF3 [Morus notabilis] |
| 8 |
Hb_004531_080 |
0.1719132841 |
- |
- |
hypothetical protein RCOM_1491960 [Ricinus communis] |
| 9 |
Hb_000638_240 |
0.1745568391 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0234100-like [Prunus mume] |
| 10 |
Hb_003540_120 |
0.1745568991 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 11 |
Hb_000077_110 |
0.1749172749 |
- |
- |
PREDICTED: uncharacterized protein LOC105646935 [Jatropha curcas] |
| 12 |
Hb_004545_040 |
0.1765519774 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2-like [Jatropha curcas] |
| 13 |
Hb_000260_380 |
0.1778957515 |
- |
- |
PREDICTED: cell division control protein 2 homolog C [Jatropha curcas] |
| 14 |
Hb_000320_310 |
0.1785285827 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C [Jatropha curcas] |
| 15 |
Hb_002871_130 |
0.1811612849 |
- |
- |
PREDICTED: non-structural maintenance of chromosomes element 4 homolog A isoform X2 [Jatropha curcas] |
| 16 |
Hb_001268_160 |
0.1824003863 |
- |
- |
Glycerophosphodiester phosphodiesterase [Medicago truncatula] |
| 17 |
Hb_001123_310 |
0.1836938528 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 18 |
Hb_020805_040 |
0.1872093803 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1A [Jatropha curcas] |
| 19 |
Hb_002284_260 |
0.1890258845 |
- |
- |
brca1 interacting protein helicase 1 brip1, putative [Ricinus communis] |
| 20 |
Hb_001619_100 |
0.1898771543 |
- |
- |
PREDICTED: uncharacterized protein LOC105630593 [Jatropha curcas] |