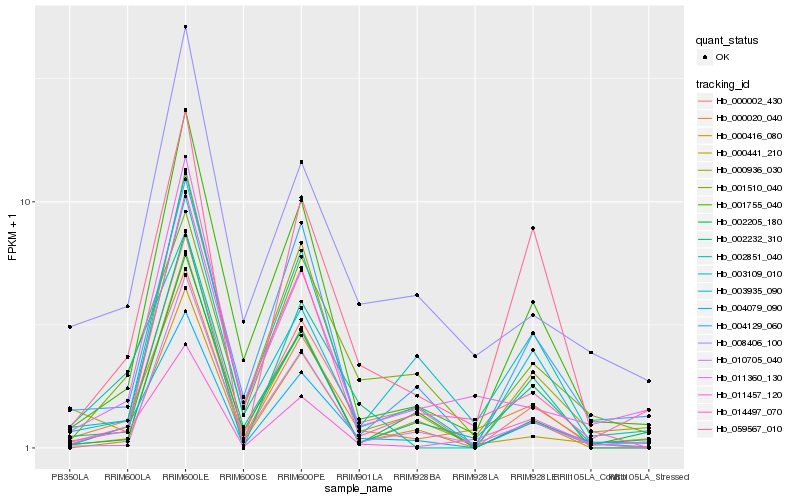
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004079_090 |
0.0 |
- |
- |
hypothetical protein RCOM_1053120 [Ricinus communis] |
| 2 |
Hb_000020_040 |
0.1309160531 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 3 |
Hb_000002_430 |
0.1353496372 |
- |
- |
PREDICTED: FHA domain-containing protein At4g14490 [Populus euphratica] |
| 4 |
Hb_001755_040 |
0.1360261117 |
- |
- |
PREDICTED: cyclin-A2-4-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000936_030 |
0.1566852823 |
- |
- |
hypothetical protein RCOM_0615680 [Ricinus communis] |
| 6 |
Hb_002232_310 |
0.1570232826 |
- |
- |
PREDICTED: uncharacterized protein LOC105635987 [Jatropha curcas] |
| 7 |
Hb_003935_090 |
0.161017664 |
- |
- |
PREDICTED: uncharacterized protein At4g38062 [Jatropha curcas] |
| 8 |
Hb_004129_060 |
0.164150866 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 9 |
Hb_001510_040 |
0.1641545654 |
- |
- |
PREDICTED: 125 kDa kinesin-related protein [Jatropha curcas] |
| 10 |
Hb_010705_040 |
0.1680527001 |
- |
- |
PREDICTED: kinesin-4 [Jatropha curcas] |
| 11 |
Hb_000416_080 |
0.1707863836 |
- |
- |
Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase, putative [Ricinus communis] |
| 12 |
Hb_002205_180 |
0.1736927365 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 13 |
Hb_059567_010 |
0.1783724555 |
- |
- |
PREDICTED: transcription factor PAR2-like [Jatropha curcas] |
| 14 |
Hb_002851_040 |
0.1789692702 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 5 [Jatropha curcas] |
| 15 |
Hb_011457_120 |
0.1799791896 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 16 |
Hb_003109_010 |
0.1800704014 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 17 |
Hb_000441_210 |
0.1815209991 |
- |
- |
PREDICTED: uncharacterized protein LOC105636842 [Jatropha curcas] |
| 18 |
Hb_008406_100 |
0.1822376411 |
- |
- |
PREDICTED: protein TPX2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_011360_130 |
0.1826920079 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_014497_070 |
0.1834111296 |
- |
- |
PREDICTED: cyclin-dependent kinase B2-2 [Jatropha curcas] |