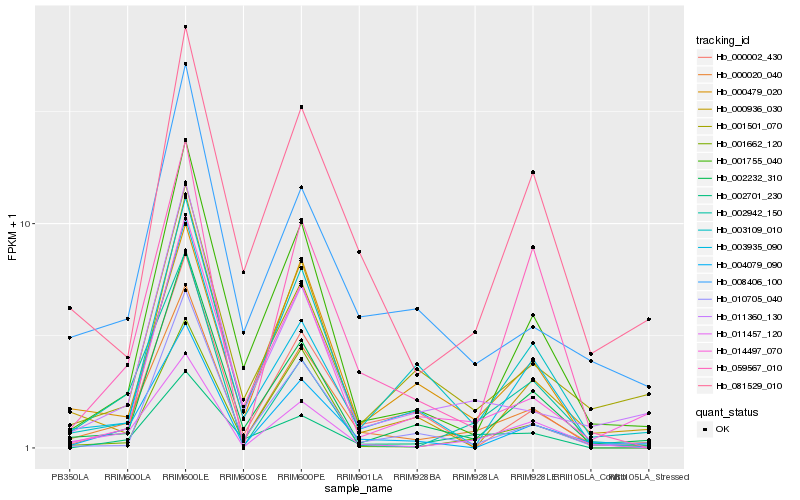
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000020_040 |
0.0 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 2 |
Hb_004079_090 |
0.1309160531 |
- |
- |
hypothetical protein RCOM_1053120 [Ricinus communis] |
| 3 |
Hb_011457_120 |
0.1309998095 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 4 |
Hb_001755_040 |
0.1342642187 |
- |
- |
PREDICTED: cyclin-A2-4-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_002232_310 |
0.1348867614 |
- |
- |
PREDICTED: uncharacterized protein LOC105635987 [Jatropha curcas] |
| 6 |
Hb_011360_130 |
0.1473993351 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_014497_070 |
0.1482726302 |
- |
- |
PREDICTED: cyclin-dependent kinase B2-2 [Jatropha curcas] |
| 8 |
Hb_001662_120 |
0.1549819562 |
- |
- |
PREDICTED: ammonium transporter 3 member 1-like [Jatropha curcas] |
| 9 |
Hb_081529_010 |
0.1552899652 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_003109_010 |
0.1582004335 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 11 |
Hb_010705_040 |
0.1610744073 |
- |
- |
PREDICTED: kinesin-4 [Jatropha curcas] |
| 12 |
Hb_008406_100 |
0.1629979496 |
- |
- |
PREDICTED: protein TPX2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_003935_090 |
0.1648164736 |
- |
- |
PREDICTED: uncharacterized protein At4g38062 [Jatropha curcas] |
| 14 |
Hb_000479_020 |
0.1653327964 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000936_030 |
0.1683022218 |
- |
- |
hypothetical protein RCOM_0615680 [Ricinus communis] |
| 16 |
Hb_001501_070 |
0.1698322025 |
- |
- |
PREDICTED: condensin complex subunit 2 [Jatropha curcas] |
| 17 |
Hb_000002_430 |
0.1716331917 |
- |
- |
PREDICTED: FHA domain-containing protein At4g14490 [Populus euphratica] |
| 18 |
Hb_002942_150 |
0.1767751179 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 19 |
Hb_059567_010 |
0.177373535 |
- |
- |
PREDICTED: transcription factor PAR2-like [Jatropha curcas] |
| 20 |
Hb_002701_230 |
0.1787044527 |
- |
- |
PREDICTED: uncharacterized protein LOC105646833 [Jatropha curcas] |