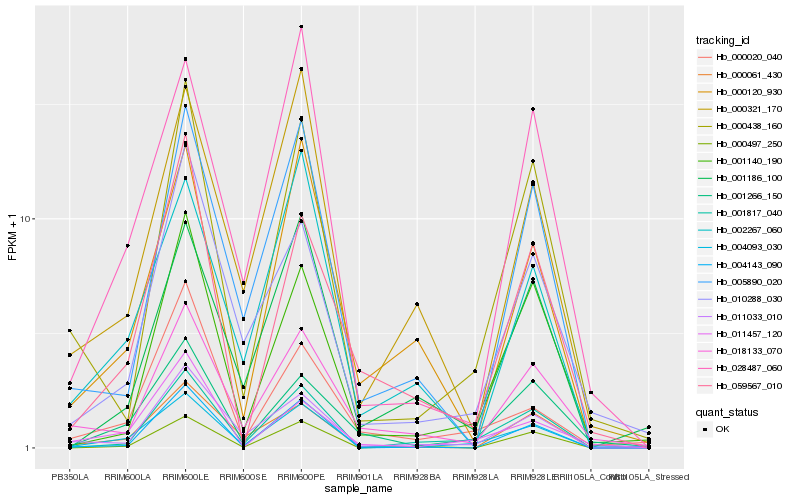
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004093_030 |
0.0 |
- |
- |
hypothetical protein RCOM_1406750 [Ricinus communis] |
| 2 |
Hb_001817_040 |
0.1454560619 |
- |
- |
myosin XI, putative [Ricinus communis] |
| 3 |
Hb_011033_010 |
0.1556690614 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 4 |
Hb_001266_150 |
0.1567688545 |
- |
- |
hypothetical protein JCGZ_14961 [Jatropha curcas] |
| 5 |
Hb_000061_430 |
0.1671010704 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_028487_060 |
0.1689672267 |
- |
- |
PREDICTED: 21 kDa protein-like [Jatropha curcas] |
| 7 |
Hb_011457_120 |
0.1713370686 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 8 |
Hb_000120_930 |
0.1729362736 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin-2-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_001140_190 |
0.1753130472 |
- |
- |
PREDICTED: A-kinase anchor protein 9 [Jatropha curcas] |
| 10 |
Hb_004143_090 |
0.1755633276 |
- |
- |
PREDICTED: VIN3-like protein 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002267_060 |
0.1773113238 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3 [Jatropha curcas] |
| 12 |
Hb_010288_030 |
0.1799778033 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 13 |
Hb_000438_160 |
0.1809601464 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001186_100 |
0.1816197162 |
- |
- |
PREDICTED: uncharacterized protein LOC105640806 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005890_020 |
0.1836728255 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 16 |
Hb_000020_040 |
0.1841804492 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 17 |
Hb_018133_070 |
0.1849110157 |
- |
- |
PREDICTED: RING-H2 finger protein ATL43 [Jatropha curcas] |
| 18 |
Hb_000321_170 |
0.1850059759 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like [Jatropha curcas] |
| 19 |
Hb_000497_250 |
0.1855674953 |
- |
- |
PREDICTED: potassium transporter 5 [Jatropha curcas] |
| 20 |
Hb_059567_010 |
0.1862865319 |
- |
- |
PREDICTED: transcription factor PAR2-like [Jatropha curcas] |