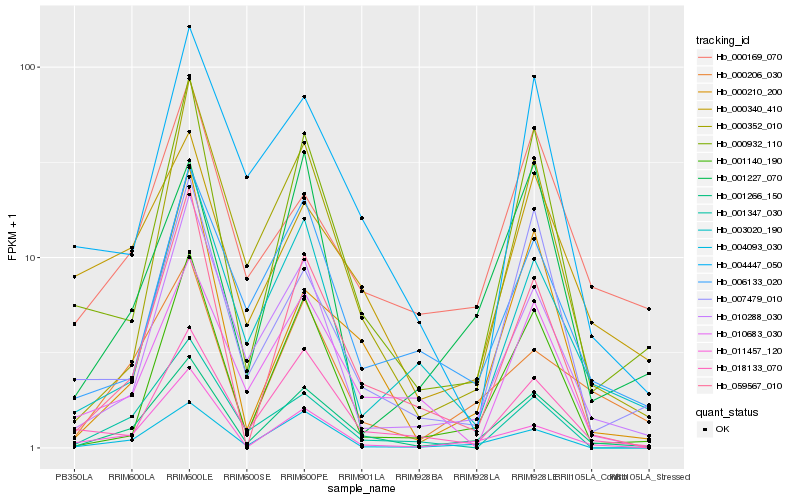
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001266_150 |
0.0 |
- |
- |
hypothetical protein JCGZ_14961 [Jatropha curcas] |
| 2 |
Hb_000932_110 |
0.146350749 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 3 |
Hb_059567_010 |
0.1539439553 |
- |
- |
PREDICTED: transcription factor PAR2-like [Jatropha curcas] |
| 4 |
Hb_004093_030 |
0.1567688545 |
- |
- |
hypothetical protein RCOM_1406750 [Ricinus communis] |
| 5 |
Hb_001140_190 |
0.1667968616 |
- |
- |
PREDICTED: A-kinase anchor protein 9 [Jatropha curcas] |
| 6 |
Hb_000352_010 |
0.1671985871 |
- |
- |
PREDICTED: nudix hydrolase 13, mitochondrial [Jatropha curcas] |
| 7 |
Hb_010288_030 |
0.1678729608 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 8 |
Hb_000206_030 |
0.1679299356 |
- |
- |
PREDICTED: uncharacterized protein LOC105639682 [Jatropha curcas] |
| 9 |
Hb_018133_070 |
0.1682145375 |
- |
- |
PREDICTED: RING-H2 finger protein ATL43 [Jatropha curcas] |
| 10 |
Hb_011457_120 |
0.1747022048 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 11 |
Hb_000210_200 |
0.1827531516 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 12 |
Hb_010683_030 |
0.1862905222 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_006133_020 |
0.1866792514 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 14 |
Hb_007479_010 |
0.1906180186 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 15 |
Hb_003020_190 |
0.1930410047 |
- |
- |
PREDICTED: methylsterol monooxygenase 2-2 [Jatropha curcas] |
| 16 |
Hb_001347_030 |
0.1942997392 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 17 |
Hb_000169_070 |
0.1957940904 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 18 |
Hb_000340_410 |
0.1973962173 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 19 |
Hb_001227_070 |
0.1980700968 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9 [Jatropha curcas] |
| 20 |
Hb_004447_050 |
0.1986375625 |
- |
- |
PREDICTED: high-light-induced protein, chloroplastic [Jatropha curcas] |