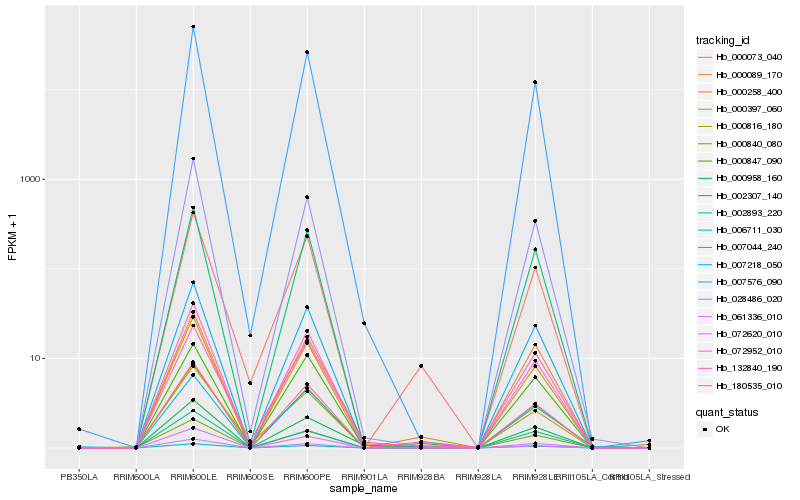
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000958_160 |
0.0 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000258_400 |
0.0967340623 |
transcription factor |
TF Family: bHLH |
transcription regulator, putative [Ricinus communis] |
| 3 |
Hb_000089_170 |
0.1022228248 |
- |
- |
PREDICTED: uncharacterized protein LOC105134595 [Populus euphratica] |
| 4 |
Hb_072952_010 |
0.1049222351 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g18430-like [Populus euphratica] |
| 5 |
Hb_007218_050 |
0.1073309904 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 6 |
Hb_000816_180 |
0.1076600963 |
- |
- |
PREDICTED: uncharacterized protein LOC105780682 [Gossypium raimondii] |
| 7 |
Hb_007576_090 |
0.1083407912 |
- |
- |
lipid transfer protein precursor [Gossypium herbaceum subsp. africanum] |
| 8 |
Hb_180535_010 |
0.1087842739 |
- |
- |
- |
| 9 |
Hb_002307_140 |
0.1110496315 |
transcription factor |
TF Family: AUX/IAA |
aux/IAA family protein [Populus trichocarpa] |
| 10 |
Hb_000073_040 |
0.1112702659 |
- |
- |
hypothetical protein POPTR_0003s21020g, partial [Populus trichocarpa] |
| 11 |
Hb_061336_010 |
0.111950006 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000847_090 |
0.1189005963 |
- |
- |
PREDICTED: uncharacterized protein LOC105644263 isoform X1 [Jatropha curcas] |
| 13 |
Hb_006711_030 |
0.1194967527 |
- |
- |
PREDICTED: rho guanine nucleotide exchange factor 8-like [Jatropha curcas] |
| 14 |
Hb_132840_190 |
0.1232097346 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_028486_020 |
0.1234798583 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000397_060 |
0.1256421208 |
- |
- |
PREDICTED: uncharacterized protein LOC105641237 [Jatropha curcas] |
| 17 |
Hb_072620_010 |
0.125762511 |
- |
- |
PREDICTED: receptor-like protein kinase 2 [Cucumis melo] |
| 18 |
Hb_002893_220 |
0.1264552185 |
- |
- |
hypothetical protein B456_001G004300 [Gossypium raimondii] |
| 19 |
Hb_000840_080 |
0.1266157434 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 9 [Jatropha curcas] |
| 20 |
Hb_007044_240 |
0.1268941125 |
- |
- |
PREDICTED: uncharacterized protein LOC105638704 isoform X1 [Jatropha curcas] |