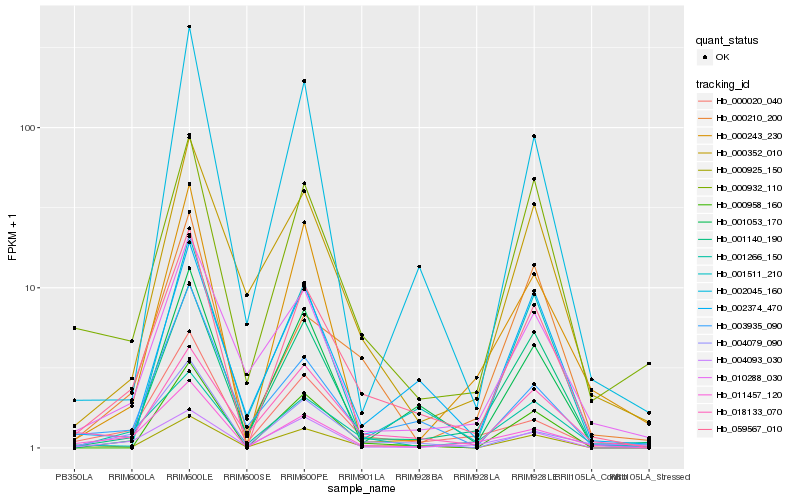
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_059567_010 |
0.0 |
- |
- |
PREDICTED: transcription factor PAR2-like [Jatropha curcas] |
| 2 |
Hb_001266_150 |
0.1539439553 |
- |
- |
hypothetical protein JCGZ_14961 [Jatropha curcas] |
| 3 |
Hb_000925_150 |
0.1558862294 |
- |
- |
PREDICTED: aluminum-activated malate transporter 8-like [Jatropha curcas] |
| 4 |
Hb_001140_190 |
0.1580889425 |
- |
- |
PREDICTED: A-kinase anchor protein 9 [Jatropha curcas] |
| 5 |
Hb_011457_120 |
0.1592143603 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 6 |
Hb_000932_110 |
0.1596629879 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 7 |
Hb_000958_160 |
0.1639840489 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_001053_170 |
0.1715059437 |
- |
- |
PREDICTED: uncharacterized protein LOC105628694 [Jatropha curcas] |
| 9 |
Hb_003935_090 |
0.171925034 |
- |
- |
PREDICTED: uncharacterized protein At4g38062 [Jatropha curcas] |
| 10 |
Hb_002374_470 |
0.1730736666 |
- |
- |
PREDICTED: filament-like plant protein isoform X2 [Jatropha curcas] |
| 11 |
Hb_000020_040 |
0.177373535 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 12 |
Hb_004079_090 |
0.1783724555 |
- |
- |
hypothetical protein RCOM_1053120 [Ricinus communis] |
| 13 |
Hb_000210_200 |
0.179894265 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 14 |
Hb_018133_070 |
0.1837019995 |
- |
- |
PREDICTED: RING-H2 finger protein ATL43 [Jatropha curcas] |
| 15 |
Hb_000352_010 |
0.1842335136 |
- |
- |
PREDICTED: nudix hydrolase 13, mitochondrial [Jatropha curcas] |
| 16 |
Hb_010288_030 |
0.1843304845 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 17 |
Hb_001511_210 |
0.1848211366 |
- |
- |
PREDICTED: protein ESKIMO 1-like [Jatropha curcas] |
| 18 |
Hb_002045_160 |
0.1850751963 |
- |
- |
PREDICTED: DJ-1 protein homolog E-like [Jatropha curcas] |
| 19 |
Hb_000243_230 |
0.1853863143 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_004093_030 |
0.1862865319 |
- |
- |
hypothetical protein RCOM_1406750 [Ricinus communis] |