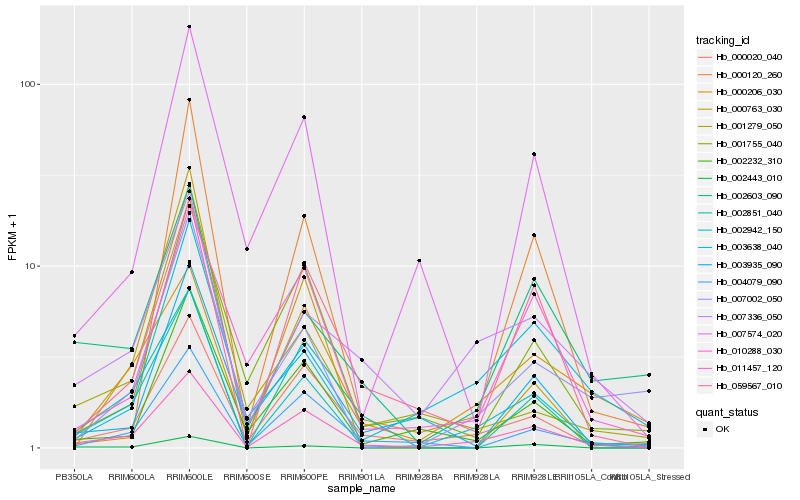
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002942_150 |
0.0 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 2 |
Hb_011457_120 |
0.1112266718 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 3 |
Hb_000020_040 |
0.1767751179 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 4 |
Hb_007002_050 |
0.1899882691 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 5 |
Hb_002443_010 |
0.1996770638 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 6 |
Hb_007336_050 |
0.2006502086 |
- |
- |
PREDICTED: uncharacterized protein LOC105630915 [Jatropha curcas] |
| 7 |
Hb_002232_310 |
0.2032592052 |
- |
- |
PREDICTED: uncharacterized protein LOC105635987 [Jatropha curcas] |
| 8 |
Hb_004079_090 |
0.2100490199 |
- |
- |
hypothetical protein RCOM_1053120 [Ricinus communis] |
| 9 |
Hb_003638_040 |
0.2140535235 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |
| 10 |
Hb_059567_010 |
0.2144656641 |
- |
- |
PREDICTED: transcription factor PAR2-like [Jatropha curcas] |
| 11 |
Hb_002603_090 |
0.2164116287 |
- |
- |
PREDICTED: GDSL esterase/lipase CPRD49-like isoform X2 [Populus euphratica] |
| 12 |
Hb_002851_040 |
0.2191348197 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 5 [Jatropha curcas] |
| 13 |
Hb_000206_030 |
0.2221717347 |
- |
- |
PREDICTED: uncharacterized protein LOC105639682 [Jatropha curcas] |
| 14 |
Hb_003935_090 |
0.2222394878 |
- |
- |
PREDICTED: uncharacterized protein At4g38062 [Jatropha curcas] |
| 15 |
Hb_000120_260 |
0.222268416 |
- |
- |
PREDICTED: ABC transporter G family member 11 [Jatropha curcas] |
| 16 |
Hb_007574_020 |
0.2237510453 |
- |
- |
PREDICTED: uncharacterized protein LOC105638146 [Jatropha curcas] |
| 17 |
Hb_001755_040 |
0.2248299658 |
- |
- |
PREDICTED: cyclin-A2-4-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000763_030 |
0.2261189148 |
- |
- |
Taxadien-5-alpha-ol O-acetyltransferase, putative [Ricinus communis] |
| 19 |
Hb_010288_030 |
0.2271759192 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 20 |
Hb_001279_050 |
0.2273633663 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |