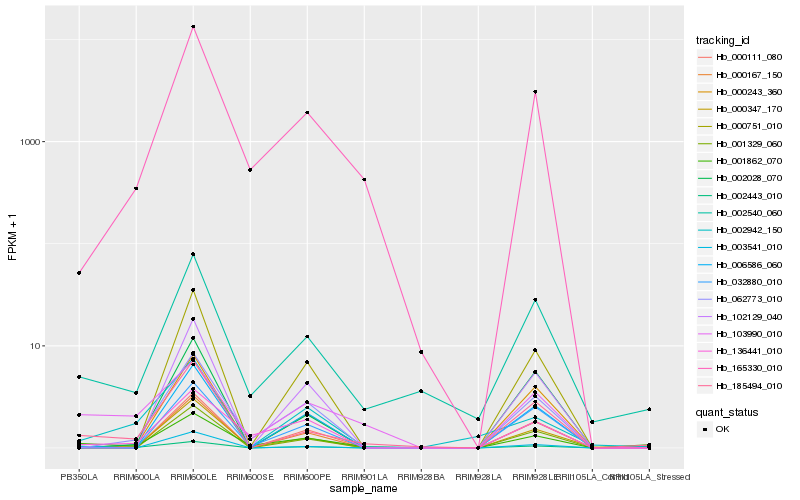
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002443_010 |
0.0 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 2 |
Hb_185494_010 |
0.1515843413 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 3 |
Hb_003541_010 |
0.1934180013 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_000347_170 |
0.1965997191 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 5 |
Hb_002942_150 |
0.1996770638 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 6 |
Hb_062773_010 |
0.2030803089 |
- |
- |
beta glucosidase [Manihot esculenta] |
| 7 |
Hb_002540_060 |
0.2075107088 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001862_070 |
0.2109517738 |
- |
- |
hypothetical protein RCOM_1155060 [Ricinus communis] |
| 9 |
Hb_000111_080 |
0.2117582258 |
- |
- |
hypothetical protein JCGZ_24534 [Jatropha curcas] |
| 10 |
Hb_000243_360 |
0.2145871113 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 isoform X1 [Vitis vinifera] |
| 11 |
Hb_165330_010 |
0.2211177496 |
- |
- |
BnaUnng00840D [Brassica napus] |
| 12 |
Hb_002028_070 |
0.2218888292 |
- |
- |
PREDICTED: circadian locomoter output cycles protein kaput [Jatropha curcas] |
| 13 |
Hb_032880_010 |
0.2222527736 |
- |
- |
PREDICTED: beta-glucosidase 24-like [Jatropha curcas] |
| 14 |
Hb_103990_010 |
0.2225012925 |
- |
- |
PREDICTED: light-induced protein, chloroplastic-like [Nicotiana sylvestris] |
| 15 |
Hb_102129_040 |
0.2234342016 |
- |
- |
PREDICTED: PTI1-like tyrosine-protein kinase 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_136441_010 |
0.2234907246 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 17 |
Hb_001329_060 |
0.2235582864 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 18 |
Hb_006586_060 |
0.2240554748 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 19 |
Hb_000167_150 |
0.2247362914 |
- |
- |
hypothetical protein POPTR_0008s03590g [Populus trichocarpa] |
| 20 |
Hb_000751_010 |
0.2249328871 |
- |
- |
conserved hypothetical protein [Ricinus communis] |