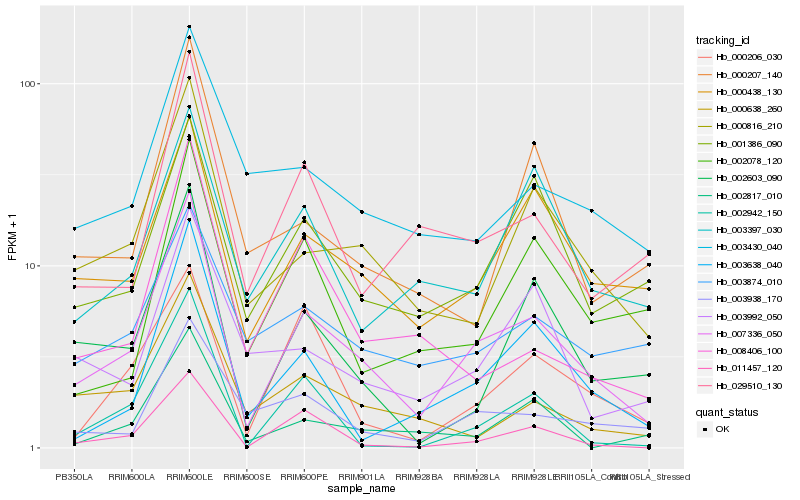
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007336_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630915 [Jatropha curcas] |
| 2 |
Hb_000816_210 |
0.1609598558 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase isoform X2 [Jatropha curcas] |
| 3 |
Hb_000438_130 |
0.1779973939 |
- |
- |
uroporphyrinogen decarboxylase, putative [Ricinus communis] |
| 4 |
Hb_002603_090 |
0.1807593797 |
- |
- |
PREDICTED: GDSL esterase/lipase CPRD49-like isoform X2 [Populus euphratica] |
| 5 |
Hb_011457_120 |
0.1865102301 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 6 |
Hb_003638_040 |
0.1875904174 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |
| 7 |
Hb_029510_130 |
0.1908788141 |
- |
- |
PREDICTED: malonate--CoA ligase [Jatropha curcas] |
| 8 |
Hb_003938_170 |
0.1944877512 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 1-like [Jatropha curcas] |
| 9 |
Hb_001386_090 |
0.1994136679 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |
| 10 |
Hb_002942_150 |
0.2006502086 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 11 |
Hb_000638_260 |
0.2008777591 |
- |
- |
hypothetical protein JCGZ_06730 [Jatropha curcas] |
| 12 |
Hb_003430_040 |
0.2061412407 |
- |
- |
spermidine synthase 1, putative [Ricinus communis] |
| 13 |
Hb_002817_010 |
0.2069595074 |
- |
- |
PREDICTED: uncharacterized protein LOC105630103 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000207_140 |
0.2073371016 |
- |
- |
hypothetical protein POPTR_0019s11310g [Populus trichocarpa] |
| 15 |
Hb_003397_030 |
0.2110391569 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 16 |
Hb_008406_100 |
0.2113010811 |
- |
- |
PREDICTED: protein TPX2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_003992_050 |
0.2116996504 |
- |
- |
delta 9 desaturase, putative [Ricinus communis] |
| 18 |
Hb_000206_030 |
0.2122249118 |
- |
- |
PREDICTED: uncharacterized protein LOC105639682 [Jatropha curcas] |
| 19 |
Hb_002078_120 |
0.2126916649 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 20 |
Hb_003874_010 |
0.213455865 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |