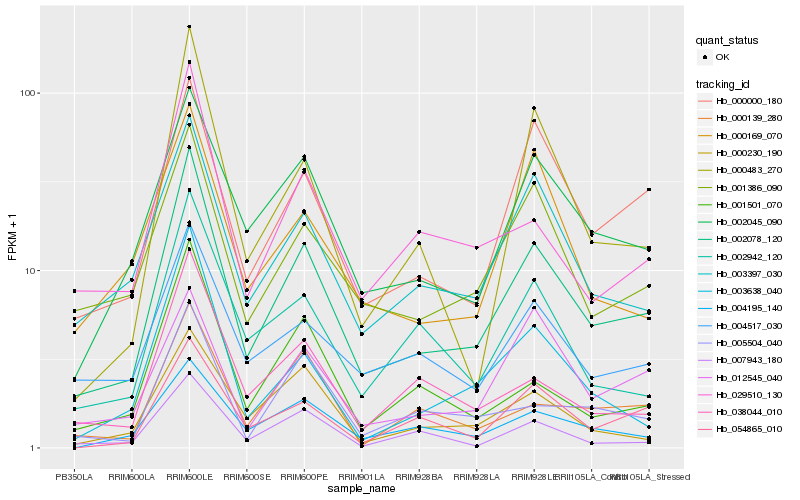
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002078_120 |
0.0 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 2 |
Hb_002045_090 |
0.1358046311 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 3 |
Hb_003397_030 |
0.138025391 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000000_180 |
0.1400859217 |
- |
- |
PREDICTED: 30S ribosomal protein S9, chloroplastic-like [Populus euphratica] |
| 5 |
Hb_001386_090 |
0.1426864454 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |
| 6 |
Hb_029510_130 |
0.1486414006 |
- |
- |
PREDICTED: malonate--CoA ligase [Jatropha curcas] |
| 7 |
Hb_000483_270 |
0.1499910055 |
- |
- |
PREDICTED: uncharacterized protein LOC105632352 [Jatropha curcas] |
| 8 |
Hb_002942_120 |
0.1512496479 |
transcription factor |
TF Family: bHLH |
Transcription factor ICE1, putative [Ricinus communis] |
| 9 |
Hb_004517_030 |
0.1548204778 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003638_040 |
0.1554937977 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |
| 11 |
Hb_038044_010 |
0.1576575342 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 12 |
Hb_004195_140 |
0.1587615883 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 13 |
Hb_001501_070 |
0.161684586 |
- |
- |
PREDICTED: condensin complex subunit 2 [Jatropha curcas] |
| 14 |
Hb_054865_010 |
0.1619298676 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860-like [Jatropha curcas] |
| 15 |
Hb_005504_040 |
0.162712479 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000230_190 |
0.1633438735 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_007943_180 |
0.1661462073 |
- |
- |
PREDICTED: uncharacterized protein LOC105637734 [Jatropha curcas] |
| 18 |
Hb_000169_070 |
0.1675865009 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 19 |
Hb_000139_280 |
0.1678168506 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase NPK1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_012545_040 |
0.1753263377 |
- |
- |
PREDICTED: uroporphyrinogen-III synthase, chloroplastic isoform X1 [Jatropha curcas] |