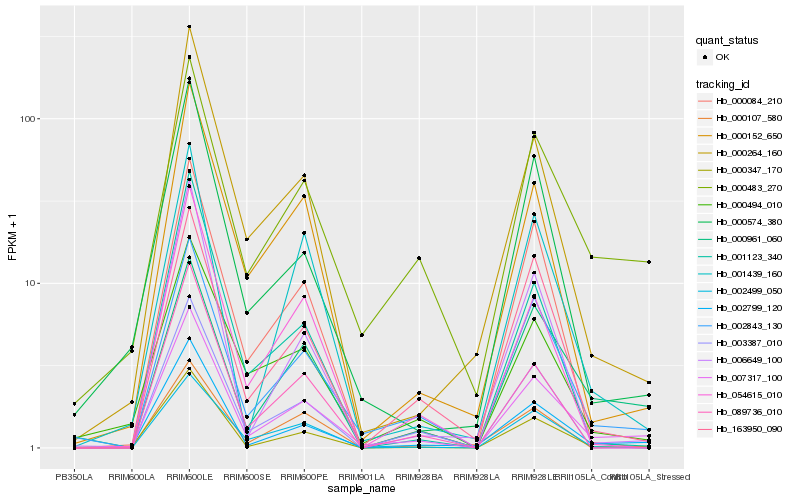
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007317_100 |
0.0 |
- |
- |
Peflin, putative [Ricinus communis] |
| 2 |
Hb_002843_130 |
0.0941718326 |
- |
- |
PREDICTED: GRF1-interacting factor 2-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_002799_120 |
0.0997823531 |
- |
- |
PREDICTED: uncharacterized protein LOC105141197 [Populus euphratica] |
| 4 |
Hb_000483_270 |
0.1253609017 |
- |
- |
PREDICTED: uncharacterized protein LOC105632352 [Jatropha curcas] |
| 5 |
Hb_000152_650 |
0.1388977713 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 6 |
Hb_000264_160 |
0.139981817 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX5-like [Jatropha curcas] |
| 7 |
Hb_000961_060 |
0.1433972259 |
- |
- |
uvb-resistance protein uvr8, putative [Ricinus communis] |
| 8 |
Hb_163950_090 |
0.1482001851 |
- |
- |
PREDICTED: traB domain-containing protein isoform X1 [Jatropha curcas] |
| 9 |
Hb_003387_010 |
0.1524203595 |
- |
- |
PREDICTED: protein kinase PINOID 2 [Gossypium raimondii] |
| 10 |
Hb_002499_050 |
0.1526453758 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 11 |
Hb_054615_010 |
0.1531808738 |
- |
- |
Chromosome-associated kinesin KIF4A, putative [Ricinus communis] |
| 12 |
Hb_000347_170 |
0.1556517357 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 13 |
Hb_089736_010 |
0.1574810432 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 7 [Populus euphratica] |
| 14 |
Hb_000574_380 |
0.1578080878 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 15 |
Hb_000084_210 |
0.1582690239 |
- |
- |
hypothetical protein POPTR_0016s08050g [Populus trichocarpa] |
| 16 |
Hb_001439_160 |
0.1582698462 |
- |
- |
PREDICTED: uncharacterized protein LOC105644097 [Jatropha curcas] |
| 17 |
Hb_001123_340 |
0.158361201 |
- |
- |
PREDICTED: uncharacterized protein LOC105123089 isoform X1 [Populus euphratica] |
| 18 |
Hb_000494_010 |
0.1584245974 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0014s04460g [Populus trichocarpa] |
| 19 |
Hb_006649_100 |
0.1592982883 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 20 |
Hb_000107_580 |
0.159376424 |
- |
- |
PREDICTED: delta(24)-sterol reductase [Jatropha curcas] |