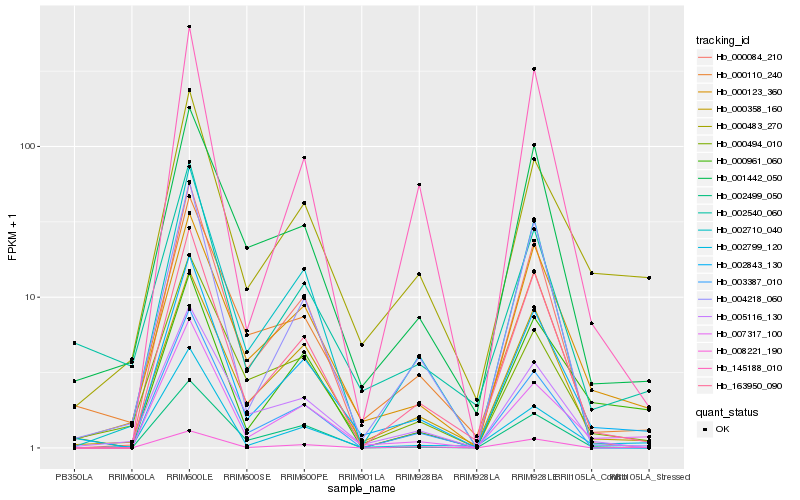
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002843_130 |
0.0 |
- |
- |
PREDICTED: GRF1-interacting factor 2-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_007317_100 |
0.0941718326 |
- |
- |
Peflin, putative [Ricinus communis] |
| 3 |
Hb_163950_090 |
0.1045919081 |
- |
- |
PREDICTED: traB domain-containing protein isoform X1 [Jatropha curcas] |
| 4 |
Hb_000358_160 |
0.1172826347 |
- |
- |
PREDICTED: uncharacterized protein LOC105637769 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000123_360 |
0.1182505761 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 1, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_000483_270 |
0.127341765 |
- |
- |
PREDICTED: uncharacterized protein LOC105632352 [Jatropha curcas] |
| 7 |
Hb_005116_130 |
0.1306188175 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa a, chloroplastic [Jatropha curcas] |
| 8 |
Hb_008221_190 |
0.132588217 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 [Jatropha curcas] |
| 9 |
Hb_000961_060 |
0.133466207 |
- |
- |
uvb-resistance protein uvr8, putative [Ricinus communis] |
| 10 |
Hb_001442_050 |
0.1376370898 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit U, chloroplastic [Jatropha curcas] |
| 11 |
Hb_002710_040 |
0.1394213091 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |
| 12 |
Hb_004218_060 |
0.1399523259 |
- |
- |
annexin [Manihot esculenta] |
| 13 |
Hb_003387_010 |
0.1417612607 |
- |
- |
PREDICTED: protein kinase PINOID 2 [Gossypium raimondii] |
| 14 |
Hb_000110_240 |
0.1424103535 |
- |
- |
PREDICTED: uncharacterized protein LOC105642006 isoform X3 [Jatropha curcas] |
| 15 |
Hb_000084_210 |
0.143286984 |
- |
- |
hypothetical protein POPTR_0016s08050g [Populus trichocarpa] |
| 16 |
Hb_002799_120 |
0.1452124871 |
- |
- |
PREDICTED: uncharacterized protein LOC105141197 [Populus euphratica] |
| 17 |
Hb_002499_050 |
0.1453227301 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 18 |
Hb_145188_010 |
0.1465770393 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 19 |
Hb_002540_060 |
0.147918374 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000494_010 |
0.1486378881 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0014s04460g [Populus trichocarpa] |