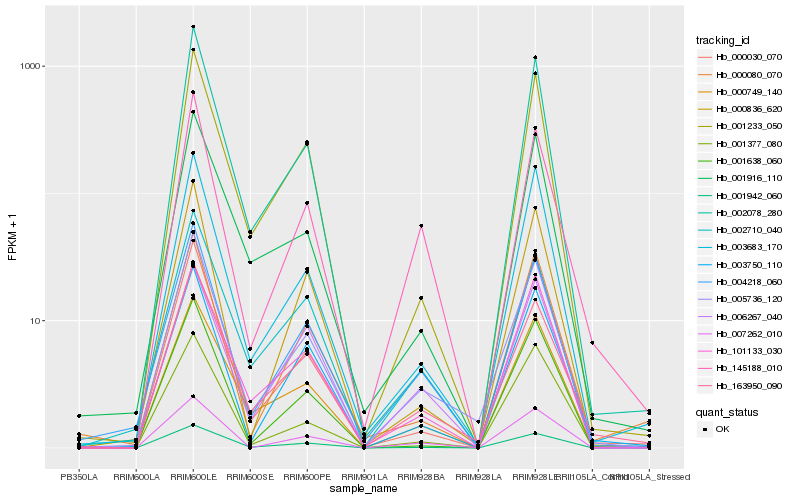
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005736_120 |
0.0 |
- |
- |
PREDICTED: carboxyvinyl-carboxyphosphonate phosphorylmutase, chloroplastic [Gossypium raimondii] |
| 2 |
Hb_163950_090 |
0.0794776883 |
- |
- |
PREDICTED: traB domain-containing protein isoform X1 [Jatropha curcas] |
| 3 |
Hb_004218_060 |
0.0822130347 |
- |
- |
annexin [Manihot esculenta] |
| 4 |
Hb_001942_060 |
0.0841516221 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000749_140 |
0.0929601766 |
- |
- |
PREDICTED: 2-succinylbenzoate--CoA ligase, chloroplastic/peroxisomal [Jatropha curcas] |
| 6 |
Hb_001233_050 |
0.0977298902 |
- |
- |
Magnesium-protoporphyrin IX monomethyl ester [oxidative] cyclase, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_003683_170 |
0.0986331978 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_101133_030 |
0.1037104435 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_145188_010 |
0.10418778 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 10 |
Hb_007262_010 |
0.1052267183 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X2 [Jatropha curcas] |
| 11 |
Hb_003750_110 |
0.1052411602 |
- |
- |
PREDICTED: aldose 1-epimerase-like [Jatropha curcas] |
| 12 |
Hb_000836_620 |
0.1087997769 |
- |
- |
PREDICTED: protochlorophyllide reductase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000030_070 |
0.1100528463 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001916_110 |
0.1113659081 |
- |
- |
PREDICTED: uncharacterized protein LOC105641510 [Jatropha curcas] |
| 15 |
Hb_002078_280 |
0.1152294057 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 16 |
Hb_002710_040 |
0.1178636656 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |
| 17 |
Hb_001377_080 |
0.1208265307 |
- |
- |
PREDICTED: uncharacterized protein LOC105642864 [Jatropha curcas] |
| 18 |
Hb_000080_070 |
0.1209772652 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP16-3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_006267_040 |
0.1213480311 |
- |
- |
PREDICTED: ABC transporter G family member 32 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001638_060 |
0.1213593549 |
- |
- |
PREDICTED: rho GTPase-activating protein 3-like isoform X1 [Populus euphratica] |