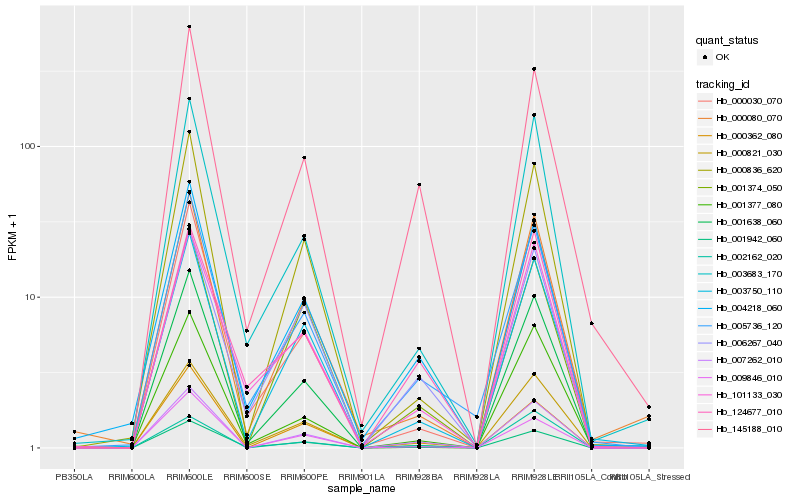
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007262_010 |
0.0 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X2 [Jatropha curcas] |
| 2 |
Hb_145188_010 |
0.0979805937 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 3 |
Hb_003750_110 |
0.0987832125 |
- |
- |
PREDICTED: aldose 1-epimerase-like [Jatropha curcas] |
| 4 |
Hb_004218_060 |
0.0990363508 |
- |
- |
annexin [Manihot esculenta] |
| 5 |
Hb_000836_620 |
0.1023332677 |
- |
- |
PREDICTED: protochlorophyllide reductase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000362_080 |
0.1043892348 |
- |
- |
hypothetical protein EUTSA_v10027329mg, partial [Eutrema salsugineum] |
| 7 |
Hb_005736_120 |
0.1052267183 |
- |
- |
PREDICTED: carboxyvinyl-carboxyphosphonate phosphorylmutase, chloroplastic [Gossypium raimondii] |
| 8 |
Hb_001374_050 |
0.1052293052 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 9 |
Hb_006267_040 |
0.1116494308 |
- |
- |
PREDICTED: ABC transporter G family member 32 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001942_060 |
0.1134751769 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003683_170 |
0.1151603055 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_009846_010 |
0.1191204267 |
- |
- |
PREDICTED: uncharacterized protein LOC105643801 [Jatropha curcas] |
| 13 |
Hb_001377_080 |
0.1200469363 |
- |
- |
PREDICTED: uncharacterized protein LOC105642864 [Jatropha curcas] |
| 14 |
Hb_000080_070 |
0.1222187787 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP16-3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001638_060 |
0.1233793508 |
- |
- |
PREDICTED: rho GTPase-activating protein 3-like isoform X1 [Populus euphratica] |
| 16 |
Hb_002162_020 |
0.1239818767 |
- |
- |
hypothetical protein JCGZ_21655 [Jatropha curcas] |
| 17 |
Hb_000821_030 |
0.1253383646 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-2 [Jatropha curcas] |
| 18 |
Hb_000030_070 |
0.1254124123 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_101133_030 |
0.1261476903 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_124677_010 |
0.1277319844 |
- |
- |
conserved hypothetical protein [Ricinus communis] |