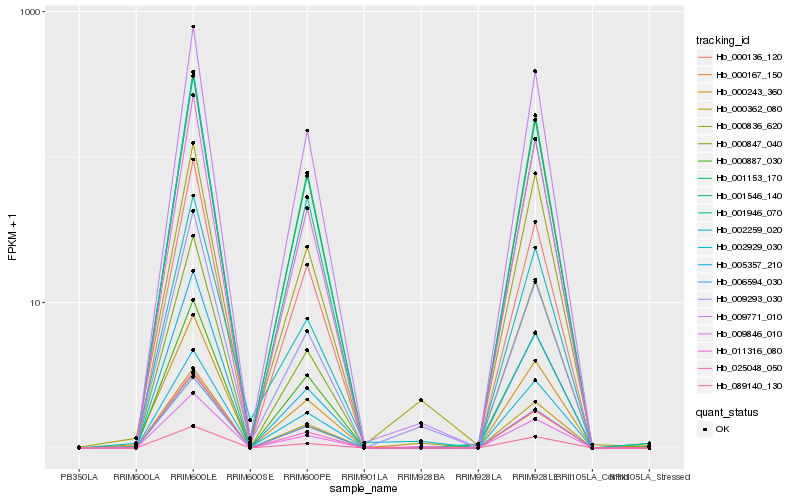
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009846_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643801 [Jatropha curcas] |
| 2 |
Hb_000362_080 |
0.0391005468 |
- |
- |
hypothetical protein EUTSA_v10027329mg, partial [Eutrema salsugineum] |
| 3 |
Hb_009293_030 |
0.0647597508 |
- |
- |
PREDICTED: salicylate carboxymethyltransferase [Jatropha curcas] |
| 4 |
Hb_009771_010 |
0.0707783185 |
- |
- |
PREDICTED: glycerol-3-phosphate 2-O-acyltransferase 6 [Populus euphratica] |
| 5 |
Hb_089140_130 |
0.0718714068 |
- |
- |
hypothetical protein POPTR_0006s14510g [Populus trichocarpa] |
| 6 |
Hb_002929_030 |
0.0719625137 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 7 |
Hb_001153_170 |
0.0740438556 |
- |
- |
PREDICTED: protein HOTHEAD [Populus euphratica] |
| 8 |
Hb_006594_030 |
0.0754795148 |
- |
- |
- |
| 9 |
Hb_011316_080 |
0.0755665801 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 10 |
Hb_000136_120 |
0.0765747697 |
- |
- |
PREDICTED: probable strigolactone esterase D14 homolog [Jatropha curcas] |
| 11 |
Hb_000847_040 |
0.0769389821 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 9 [Jatropha curcas] |
| 12 |
Hb_000167_150 |
0.077813298 |
- |
- |
hypothetical protein POPTR_0008s03590g [Populus trichocarpa] |
| 13 |
Hb_001946_070 |
0.0783436899 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_025048_050 |
0.0785341537 |
- |
- |
PREDICTED: protein PHYTOCHROME KINASE SUBSTRATE 3 [Jatropha curcas] |
| 15 |
Hb_000836_620 |
0.0818036958 |
- |
- |
PREDICTED: protochlorophyllide reductase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002259_020 |
0.0861917632 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |
| 17 |
Hb_001546_140 |
0.0872690458 |
- |
- |
Auxin-binding protein ABP19a precursor, putative [Ricinus communis] |
| 18 |
Hb_005357_210 |
0.0913161277 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein MERISTEM L1 [Jatropha curcas] |
| 19 |
Hb_000243_360 |
0.0914568265 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 isoform X1 [Vitis vinifera] |
| 20 |
Hb_000887_030 |
0.0920416577 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |