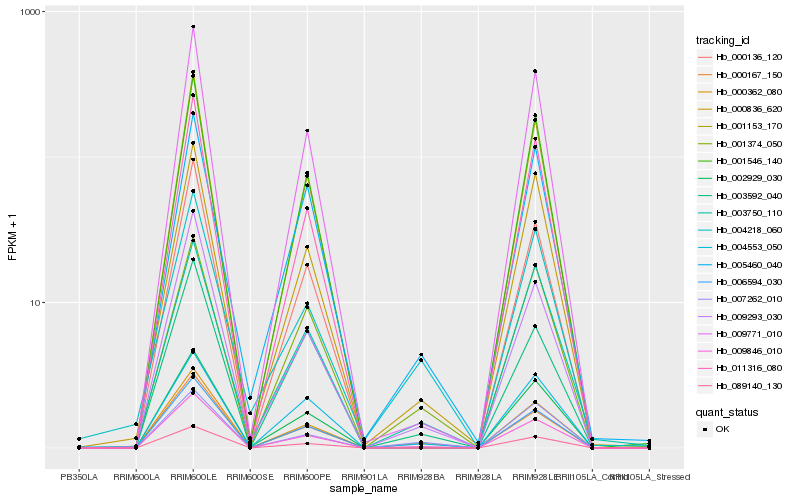
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000362_080 |
0.0 |
- |
- |
hypothetical protein EUTSA_v10027329mg, partial [Eutrema salsugineum] |
| 2 |
Hb_009846_010 |
0.0391005468 |
- |
- |
PREDICTED: uncharacterized protein LOC105643801 [Jatropha curcas] |
| 3 |
Hb_009293_030 |
0.0852948165 |
- |
- |
PREDICTED: salicylate carboxymethyltransferase [Jatropha curcas] |
| 4 |
Hb_001374_050 |
0.0894564929 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 5 |
Hb_002929_030 |
0.0925303777 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 6 |
Hb_000836_620 |
0.0952091704 |
- |
- |
PREDICTED: protochlorophyllide reductase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_009771_010 |
0.0961511382 |
- |
- |
PREDICTED: glycerol-3-phosphate 2-O-acyltransferase 6 [Populus euphratica] |
| 8 |
Hb_006594_030 |
0.0992874547 |
- |
- |
- |
| 9 |
Hb_001153_170 |
0.0996231449 |
- |
- |
PREDICTED: protein HOTHEAD [Populus euphratica] |
| 10 |
Hb_089140_130 |
0.0997519106 |
- |
- |
hypothetical protein POPTR_0006s14510g [Populus trichocarpa] |
| 11 |
Hb_000136_120 |
0.1021978637 |
- |
- |
PREDICTED: probable strigolactone esterase D14 homolog [Jatropha curcas] |
| 12 |
Hb_000167_150 |
0.1029359193 |
- |
- |
hypothetical protein POPTR_0008s03590g [Populus trichocarpa] |
| 13 |
Hb_003592_040 |
0.1038051179 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 9 [Jatropha curcas] |
| 14 |
Hb_011316_080 |
0.1039826498 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 15 |
Hb_007262_010 |
0.1043892348 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X2 [Jatropha curcas] |
| 16 |
Hb_003750_110 |
0.1060930557 |
- |
- |
PREDICTED: aldose 1-epimerase-like [Jatropha curcas] |
| 17 |
Hb_004218_060 |
0.1066091954 |
- |
- |
annexin [Manihot esculenta] |
| 18 |
Hb_005460_040 |
0.1069202756 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 2 [Jatropha curcas] |
| 19 |
Hb_001546_140 |
0.1076991086 |
- |
- |
Auxin-binding protein ABP19a precursor, putative [Ricinus communis] |
| 20 |
Hb_004553_050 |
0.1083275059 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g10020 [Jatropha curcas] |