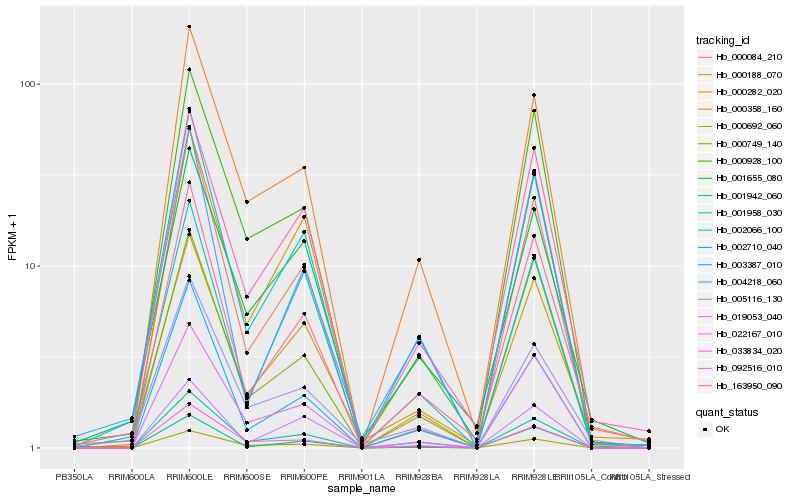
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002710_040 |
0.0 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |
| 2 |
Hb_001958_030 |
0.0725958513 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 3 |
Hb_003387_010 |
0.0880158272 |
- |
- |
PREDICTED: protein kinase PINOID 2 [Gossypium raimondii] |
| 4 |
Hb_000188_070 |
0.090206878 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 5 |
Hb_163950_090 |
0.0931699118 |
- |
- |
PREDICTED: traB domain-containing protein isoform X1 [Jatropha curcas] |
| 6 |
Hb_004218_060 |
0.0936927792 |
- |
- |
annexin [Manihot esculenta] |
| 7 |
Hb_001942_060 |
0.0951458474 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000084_210 |
0.0963991545 |
- |
- |
hypothetical protein POPTR_0016s08050g [Populus trichocarpa] |
| 9 |
Hb_000692_060 |
0.0992676467 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 10 |
Hb_092516_010 |
0.0992867018 |
- |
- |
hypothetical protein JCGZ_12248 [Jatropha curcas] |
| 11 |
Hb_022167_010 |
0.09971656 |
- |
- |
Kinase, putative isoform 2 [Theobroma cacao] |
| 12 |
Hb_033834_020 |
0.1008268482 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000358_160 |
0.1035670446 |
- |
- |
PREDICTED: uncharacterized protein LOC105637769 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002066_100 |
0.1042380249 |
- |
- |
PREDICTED: protein disulfide-isomerase SCO2 [Jatropha curcas] |
| 15 |
Hb_019053_040 |
0.1043534546 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_000749_140 |
0.107132615 |
- |
- |
PREDICTED: 2-succinylbenzoate--CoA ligase, chloroplastic/peroxisomal [Jatropha curcas] |
| 17 |
Hb_000928_100 |
0.10896927 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 18 |
Hb_001655_080 |
0.1096673485 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 19 |
Hb_000282_020 |
0.1125338395 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005116_130 |
0.1135491623 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa a, chloroplastic [Jatropha curcas] |