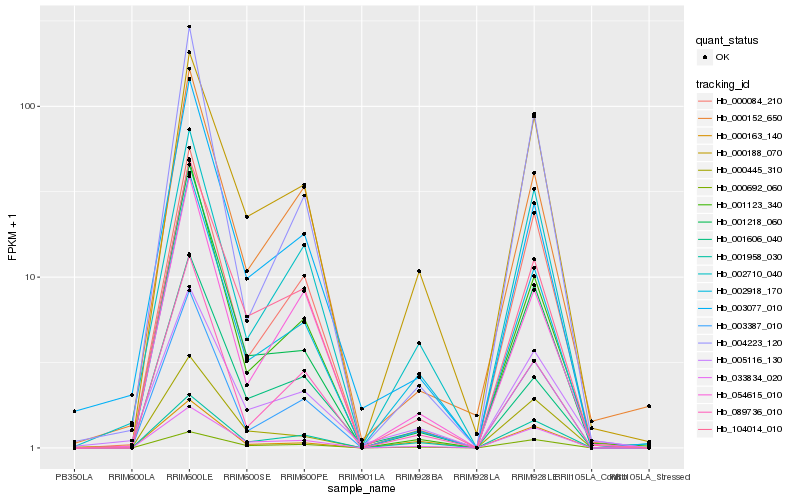
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002918_170 |
0.0 |
- |
- |
PREDICTED: MACPF domain-containing protein NSL1 [Jatropha curcas] |
| 2 |
Hb_003387_010 |
0.0508764514 |
- |
- |
PREDICTED: protein kinase PINOID 2 [Gossypium raimondii] |
| 3 |
Hb_001958_030 |
0.0985433229 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 4 |
Hb_000188_070 |
0.1000055515 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 5 |
Hb_089736_010 |
0.1017174943 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 7 [Populus euphratica] |
| 6 |
Hb_000163_140 |
0.1023677961 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 7 |
Hb_104014_010 |
0.104149973 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39020 isoform X2 [Jatropha curcas] |
| 8 |
Hb_033834_020 |
0.1046331005 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_054615_010 |
0.1134492899 |
- |
- |
Chromosome-associated kinesin KIF4A, putative [Ricinus communis] |
| 10 |
Hb_002710_040 |
0.1155828706 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |
| 11 |
Hb_003077_010 |
0.1170273333 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit O, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_001123_340 |
0.1187286508 |
- |
- |
PREDICTED: uncharacterized protein LOC105123089 isoform X1 [Populus euphratica] |
| 13 |
Hb_001218_060 |
0.1194101643 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005116_130 |
0.1208496889 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa a, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000445_310 |
0.1220904628 |
- |
- |
hypothetical protein B456_011G218000 [Gossypium raimondii] |
| 16 |
Hb_001606_040 |
0.128455109 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 17 |
Hb_004223_120 |
0.1292123003 |
- |
- |
PREDICTED: protein SPA, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000152_650 |
0.1312439418 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 19 |
Hb_000084_210 |
0.1318161676 |
- |
- |
hypothetical protein POPTR_0016s08050g [Populus trichocarpa] |
| 20 |
Hb_000692_060 |
0.1329450091 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |