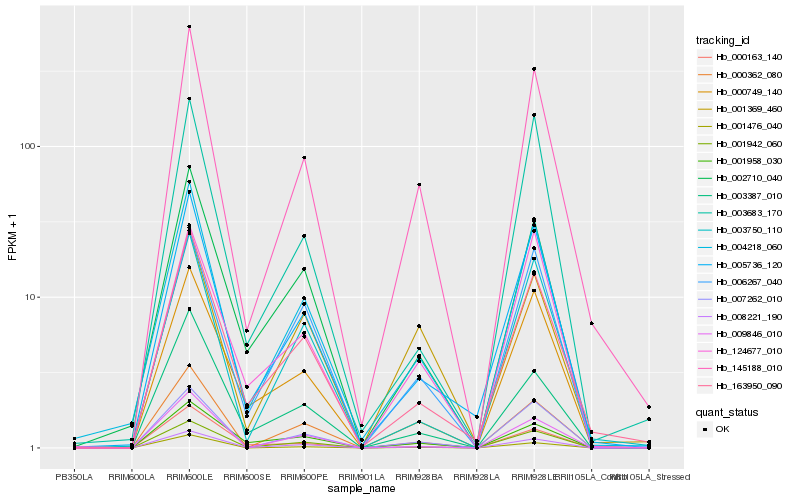
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_145188_010 |
0.0 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 2 |
Hb_004218_060 |
0.086269467 |
- |
- |
annexin [Manihot esculenta] |
| 3 |
Hb_007262_010 |
0.0979805937 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X2 [Jatropha curcas] |
| 4 |
Hb_163950_090 |
0.1009055922 |
- |
- |
PREDICTED: traB domain-containing protein isoform X1 [Jatropha curcas] |
| 5 |
Hb_005736_120 |
0.10418778 |
- |
- |
PREDICTED: carboxyvinyl-carboxyphosphonate phosphorylmutase, chloroplastic [Gossypium raimondii] |
| 6 |
Hb_001476_040 |
0.1049847543 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 7 |
Hb_000362_080 |
0.1201008691 |
- |
- |
hypothetical protein EUTSA_v10027329mg, partial [Eutrema salsugineum] |
| 8 |
Hb_008221_190 |
0.1227926443 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 [Jatropha curcas] |
| 9 |
Hb_001369_460 |
0.1263694958 |
- |
- |
vacuolar invertase [Manihot esculenta] |
| 10 |
Hb_006267_040 |
0.1275355443 |
- |
- |
PREDICTED: ABC transporter G family member 32 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003750_110 |
0.128057775 |
- |
- |
PREDICTED: aldose 1-epimerase-like [Jatropha curcas] |
| 12 |
Hb_001942_060 |
0.1296317975 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002710_040 |
0.1346768025 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |
| 14 |
Hb_000749_140 |
0.1359647453 |
- |
- |
PREDICTED: 2-succinylbenzoate--CoA ligase, chloroplastic/peroxisomal [Jatropha curcas] |
| 15 |
Hb_000163_140 |
0.136701927 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 16 |
Hb_124677_010 |
0.1390825541 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_009846_010 |
0.1399851722 |
- |
- |
PREDICTED: uncharacterized protein LOC105643801 [Jatropha curcas] |
| 18 |
Hb_001958_030 |
0.1441871395 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 19 |
Hb_003387_010 |
0.1452463058 |
- |
- |
PREDICTED: protein kinase PINOID 2 [Gossypium raimondii] |
| 20 |
Hb_003683_170 |
0.1465645671 |
- |
- |
conserved hypothetical protein [Ricinus communis] |