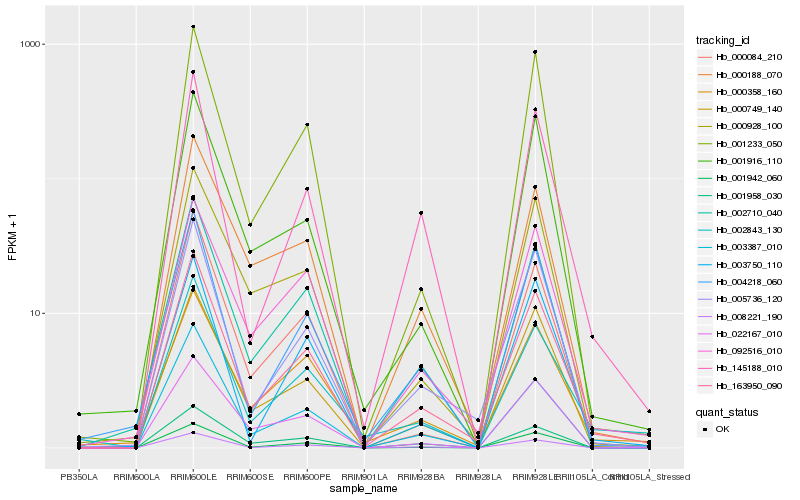
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_163950_090 |
0.0 |
- |
- |
PREDICTED: traB domain-containing protein isoform X1 [Jatropha curcas] |
| 2 |
Hb_005736_120 |
0.0794776883 |
- |
- |
PREDICTED: carboxyvinyl-carboxyphosphonate phosphorylmutase, chloroplastic [Gossypium raimondii] |
| 3 |
Hb_001942_060 |
0.0886502569 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002710_040 |
0.0931699118 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |
| 5 |
Hb_000749_140 |
0.0974928082 |
- |
- |
PREDICTED: 2-succinylbenzoate--CoA ligase, chloroplastic/peroxisomal [Jatropha curcas] |
| 6 |
Hb_004218_060 |
0.0994155008 |
- |
- |
annexin [Manihot esculenta] |
| 7 |
Hb_145188_010 |
0.1009055922 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 8 |
Hb_092516_010 |
0.1015437718 |
- |
- |
hypothetical protein JCGZ_12248 [Jatropha curcas] |
| 9 |
Hb_001233_050 |
0.1028147291 |
- |
- |
Magnesium-protoporphyrin IX monomethyl ester [oxidative] cyclase, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_002843_130 |
0.1045919081 |
- |
- |
PREDICTED: GRF1-interacting factor 2-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_000084_210 |
0.1046395575 |
- |
- |
hypothetical protein POPTR_0016s08050g [Populus trichocarpa] |
| 12 |
Hb_000358_160 |
0.1055146685 |
- |
- |
PREDICTED: uncharacterized protein LOC105637769 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000188_070 |
0.1106354239 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 14 |
Hb_003387_010 |
0.1116067429 |
- |
- |
PREDICTED: protein kinase PINOID 2 [Gossypium raimondii] |
| 15 |
Hb_001958_030 |
0.1127380447 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 16 |
Hb_008221_190 |
0.1149126937 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 [Jatropha curcas] |
| 17 |
Hb_003750_110 |
0.1154932627 |
- |
- |
PREDICTED: aldose 1-epimerase-like [Jatropha curcas] |
| 18 |
Hb_022167_010 |
0.1156812894 |
- |
- |
Kinase, putative isoform 2 [Theobroma cacao] |
| 19 |
Hb_001916_110 |
0.1157049082 |
- |
- |
PREDICTED: uncharacterized protein LOC105641510 [Jatropha curcas] |
| 20 |
Hb_000928_100 |
0.1179322315 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |