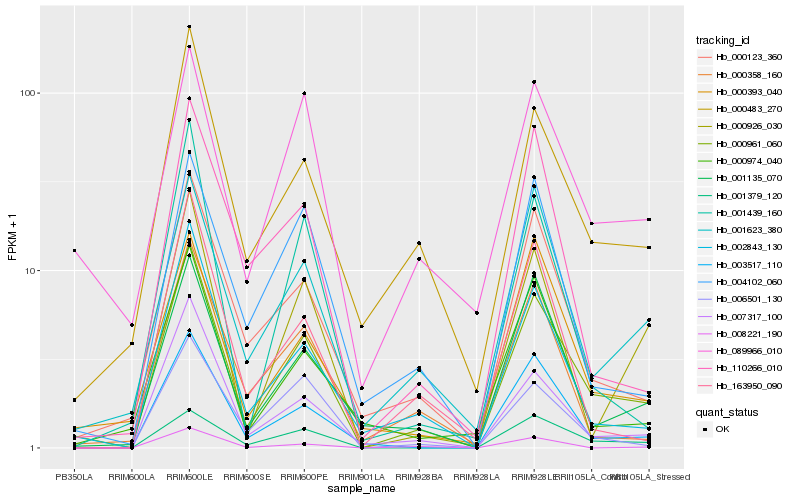
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000961_060 |
0.0 |
- |
- |
uvb-resistance protein uvr8, putative [Ricinus communis] |
| 2 |
Hb_002843_130 |
0.133466207 |
- |
- |
PREDICTED: GRF1-interacting factor 2-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_000483_270 |
0.1429615511 |
- |
- |
PREDICTED: uncharacterized protein LOC105632352 [Jatropha curcas] |
| 4 |
Hb_007317_100 |
0.1433972259 |
- |
- |
Peflin, putative [Ricinus communis] |
| 5 |
Hb_000123_360 |
0.1461775057 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 1, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_003517_110 |
0.1479626473 |
- |
- |
PREDICTED: uncharacterized protein LOC105633058 [Jatropha curcas] |
| 7 |
Hb_001135_070 |
0.1542069398 |
- |
- |
glutathione-s-transferase omega, putative [Ricinus communis] |
| 8 |
Hb_001379_120 |
0.1571022659 |
- |
- |
hypothetical protein POPTR_0001s29730g [Populus trichocarpa] |
| 9 |
Hb_001623_380 |
0.161436089 |
- |
- |
PREDICTED: 7-hydroxymethyl chlorophyll a reductase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000926_030 |
0.1693388074 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_006501_130 |
0.1734167349 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000358_160 |
0.1736591282 |
- |
- |
PREDICTED: uncharacterized protein LOC105637769 isoform X1 [Jatropha curcas] |
| 13 |
Hb_008221_190 |
0.1753744808 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 [Jatropha curcas] |
| 14 |
Hb_000393_040 |
0.1754222468 |
- |
- |
PREDICTED: uncharacterized protein LOC105128208 [Populus euphratica] |
| 15 |
Hb_004102_060 |
0.1767691603 |
- |
- |
JHL25H03.11 [Jatropha curcas] |
| 16 |
Hb_089966_010 |
0.1768302272 |
- |
- |
- |
| 17 |
Hb_001439_160 |
0.1771508015 |
- |
- |
PREDICTED: uncharacterized protein LOC105644097 [Jatropha curcas] |
| 18 |
Hb_163950_090 |
0.1773385342 |
- |
- |
PREDICTED: traB domain-containing protein isoform X1 [Jatropha curcas] |
| 19 |
Hb_000974_040 |
0.1778204591 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g49770 [Jatropha curcas] |
| 20 |
Hb_110266_010 |
0.1790441976 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g21222-like [Jatropha curcas] |