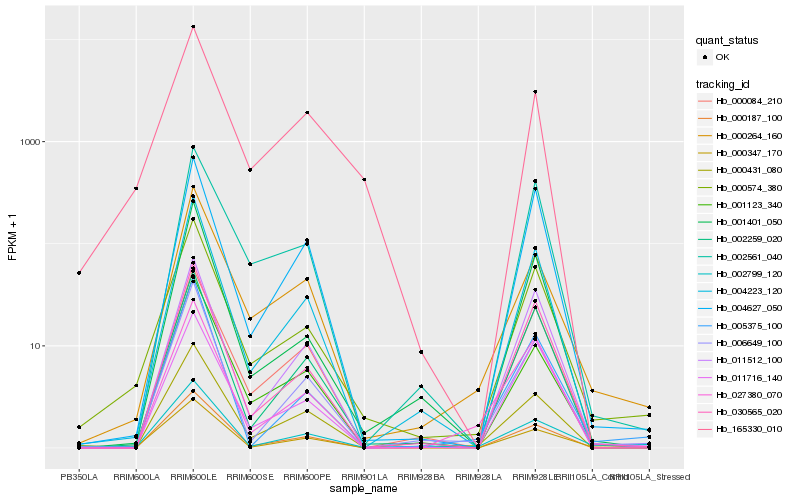
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000574_380 |
0.0 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 2 |
Hb_004223_120 |
0.1162820198 |
- |
- |
PREDICTED: protein SPA, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000347_170 |
0.1166648209 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 4 |
Hb_030565_020 |
0.1202475026 |
- |
- |
PREDICTED: protein ECERIFERUM 26 [Jatropha curcas] |
| 5 |
Hb_000264_160 |
0.1228220926 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX5-like [Jatropha curcas] |
| 6 |
Hb_000431_080 |
0.1247134766 |
- |
- |
PREDICTED: uncharacterized protein LOC105632547 [Jatropha curcas] |
| 7 |
Hb_001401_050 |
0.1270013392 |
- |
- |
PREDICTED: chlorophyll a-b binding protein P4, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002259_020 |
0.130317116 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |
| 9 |
Hb_002799_120 |
0.1304733452 |
- |
- |
PREDICTED: uncharacterized protein LOC105141197 [Populus euphratica] |
| 10 |
Hb_006649_100 |
0.1331496555 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 11 |
Hb_000084_210 |
0.1332292883 |
- |
- |
hypothetical protein POPTR_0016s08050g [Populus trichocarpa] |
| 12 |
Hb_002561_040 |
0.1336309041 |
- |
- |
Photosynthetic electron transfer C [Theobroma cacao] |
| 13 |
Hb_000187_100 |
0.1351316724 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 14 |
Hb_011716_140 |
0.1374792057 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein J [Jatropha curcas] |
| 15 |
Hb_165330_010 |
0.1375686892 |
- |
- |
BnaUnng00840D [Brassica napus] |
| 16 |
Hb_005375_100 |
0.1421654633 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH57-like [Jatropha curcas] |
| 17 |
Hb_011512_100 |
0.1425160576 |
- |
- |
PREDICTED: uncharacterized protein LOC105650382 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004627_050 |
0.1433681169 |
- |
- |
Chlorophyll a-b binding protein 6A [Morus notabilis] |
| 19 |
Hb_001123_340 |
0.1435217303 |
- |
- |
PREDICTED: uncharacterized protein LOC105123089 isoform X1 [Populus euphratica] |
| 20 |
Hb_027380_070 |
0.1468384686 |
- |
- |
conserved hypothetical protein [Ricinus communis] |