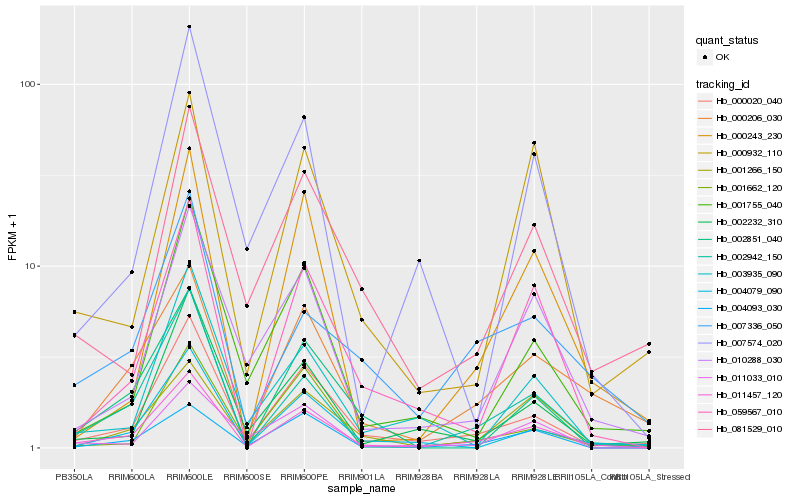
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011457_120 |
0.0 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 2 |
Hb_002942_150 |
0.1112266718 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 3 |
Hb_000020_040 |
0.1309998095 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 4 |
Hb_059567_010 |
0.1592143603 |
- |
- |
PREDICTED: transcription factor PAR2-like [Jatropha curcas] |
| 5 |
Hb_004093_030 |
0.1713370686 |
- |
- |
hypothetical protein RCOM_1406750 [Ricinus communis] |
| 6 |
Hb_000206_030 |
0.1713627654 |
- |
- |
PREDICTED: uncharacterized protein LOC105639682 [Jatropha curcas] |
| 7 |
Hb_001266_150 |
0.1747022048 |
- |
- |
hypothetical protein JCGZ_14961 [Jatropha curcas] |
| 8 |
Hb_004079_090 |
0.1799791896 |
- |
- |
hypothetical protein RCOM_1053120 [Ricinus communis] |
| 9 |
Hb_002851_040 |
0.186377205 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 5 [Jatropha curcas] |
| 10 |
Hb_007336_050 |
0.1865102301 |
- |
- |
PREDICTED: uncharacterized protein LOC105630915 [Jatropha curcas] |
| 11 |
Hb_010288_030 |
0.1880454614 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 12 |
Hb_000243_230 |
0.1891175661 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_002232_310 |
0.1939337397 |
- |
- |
PREDICTED: uncharacterized protein LOC105635987 [Jatropha curcas] |
| 14 |
Hb_001755_040 |
0.1963774132 |
- |
- |
PREDICTED: cyclin-A2-4-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_003935_090 |
0.1991384152 |
- |
- |
PREDICTED: uncharacterized protein At4g38062 [Jatropha curcas] |
| 16 |
Hb_081529_010 |
0.2080982409 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_011033_010 |
0.2084646589 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 18 |
Hb_007574_020 |
0.2088312484 |
- |
- |
PREDICTED: uncharacterized protein LOC105638146 [Jatropha curcas] |
| 19 |
Hb_000932_110 |
0.2103154081 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 20 |
Hb_001662_120 |
0.2116384989 |
- |
- |
PREDICTED: ammonium transporter 3 member 1-like [Jatropha curcas] |