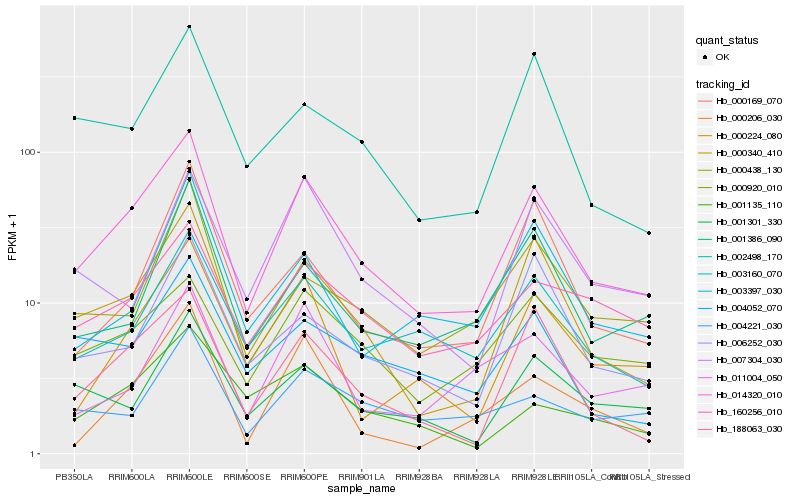
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014320_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629932 [Jatropha curcas] |
| 2 |
Hb_000340_410 |
0.1131099424 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 3 |
Hb_188063_030 |
0.1378316689 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 4 |
Hb_003160_070 |
0.141605597 |
- |
- |
PREDICTED: VIN3-like protein 1 [Jatropha curcas] |
| 5 |
Hb_000224_080 |
0.1491745809 |
- |
- |
hypothetical protein RCOM_0510880 [Ricinus communis] |
| 6 |
Hb_160256_010 |
0.1541044644 |
- |
- |
- |
| 7 |
Hb_000920_010 |
0.1546521789 |
- |
- |
hypothetical protein PRUPE_ppa013940mg [Prunus persica] |
| 8 |
Hb_001301_330 |
0.1628980386 |
- |
- |
OTU-like cysteine protease family protein [Populus trichocarpa] |
| 9 |
Hb_002498_170 |
0.1635888262 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 10 |
Hb_001386_090 |
0.1644163168 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |
| 11 |
Hb_000438_130 |
0.1644358802 |
- |
- |
uroporphyrinogen decarboxylase, putative [Ricinus communis] |
| 12 |
Hb_004052_070 |
0.1659751012 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 13 |
Hb_003397_030 |
0.1703974182 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_004221_030 |
0.1710068113 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_006252_030 |
0.1721083732 |
- |
- |
PREDICTED: uncharacterized protein LOC105638333 [Jatropha curcas] |
| 16 |
Hb_000169_070 |
0.1749310941 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 17 |
Hb_007304_030 |
0.176395472 |
- |
- |
PREDICTED: uncharacterized protein LOC105635926 isoform X2 [Jatropha curcas] |
| 18 |
Hb_011004_050 |
0.1765590234 |
- |
- |
PREDICTED: kinesin-13A [Jatropha curcas] |
| 19 |
Hb_000206_030 |
0.1778165848 |
- |
- |
PREDICTED: uncharacterized protein LOC105639682 [Jatropha curcas] |
| 20 |
Hb_001135_110 |
0.178226143 |
- |
- |
- |