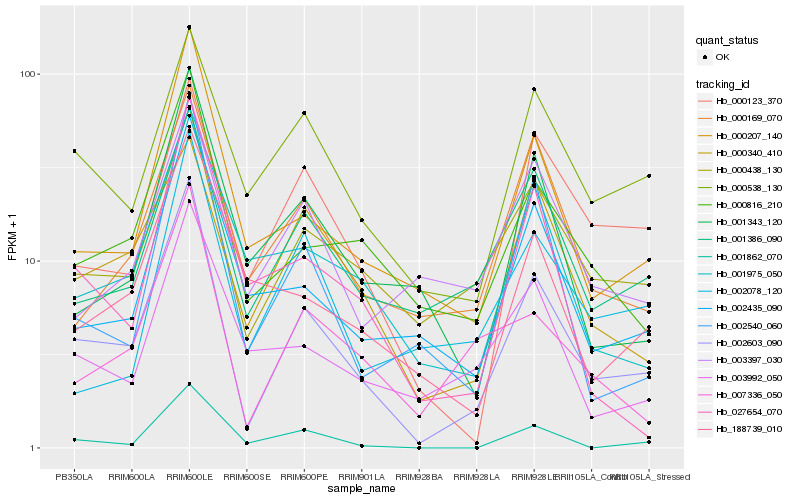
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002603_090 |
0.0 |
- |
- |
PREDICTED: GDSL esterase/lipase CPRD49-like isoform X2 [Populus euphratica] |
| 2 |
Hb_000207_140 |
0.1576815845 |
- |
- |
hypothetical protein POPTR_0019s11310g [Populus trichocarpa] |
| 3 |
Hb_000816_210 |
0.1670350891 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase isoform X2 [Jatropha curcas] |
| 4 |
Hb_000438_130 |
0.1719340404 |
- |
- |
uroporphyrinogen decarboxylase, putative [Ricinus communis] |
| 5 |
Hb_007336_050 |
0.1807593797 |
- |
- |
PREDICTED: uncharacterized protein LOC105630915 [Jatropha curcas] |
| 6 |
Hb_002435_090 |
0.1836793934 |
- |
- |
PREDICTED: membrane-associated 30 kDa protein, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002540_060 |
0.1842283831 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001862_070 |
0.1863971708 |
- |
- |
hypothetical protein RCOM_1155060 [Ricinus communis] |
| 9 |
Hb_001386_090 |
0.1872511798 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |
| 10 |
Hb_000340_410 |
0.1902672881 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 11 |
Hb_000169_070 |
0.192472129 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 12 |
Hb_001975_050 |
0.1925789185 |
- |
- |
Tyrosine-specific transport protein, putative [Ricinus communis] |
| 13 |
Hb_000123_370 |
0.1936653243 |
- |
- |
PREDICTED: psbP-like protein 1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000538_130 |
0.1951118116 |
- |
- |
phytoene synthase 2 [Manihot esculenta] |
| 15 |
Hb_027654_070 |
0.1973145741 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001343_120 |
0.1979813008 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_188739_010 |
0.1991319523 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 18 |
Hb_003992_050 |
0.2015978389 |
- |
- |
delta 9 desaturase, putative [Ricinus communis] |
| 19 |
Hb_003397_030 |
0.2087101102 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002078_120 |
0.2106039471 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |