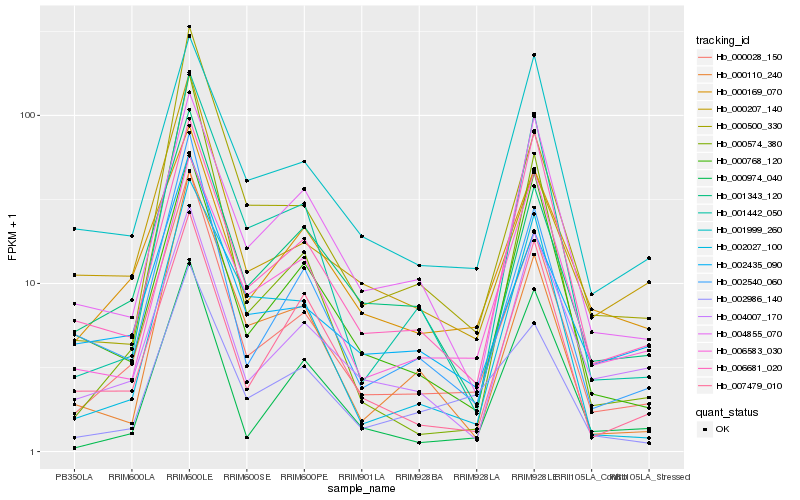
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000028_150 |
0.0 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 2 |
Hb_002540_060 |
0.1040999288 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002986_140 |
0.1220335782 |
- |
- |
PREDICTED: uncharacterized protein LOC105639684 [Jatropha curcas] |
| 4 |
Hb_001343_120 |
0.1232290484 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002435_090 |
0.1332183582 |
- |
- |
PREDICTED: membrane-associated 30 kDa protein, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001442_050 |
0.1343117603 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit U, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000500_330 |
0.1374526906 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET2a-like [Jatropha curcas] |
| 8 |
Hb_000169_070 |
0.1407670554 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 9 |
Hb_001999_260 |
0.1423328211 |
- |
- |
PREDICTED: 50S ribosomal protein L4, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000207_140 |
0.1431937703 |
- |
- |
hypothetical protein POPTR_0019s11310g [Populus trichocarpa] |
| 11 |
Hb_000974_040 |
0.1433690484 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g49770 [Jatropha curcas] |
| 12 |
Hb_000110_240 |
0.1436655483 |
- |
- |
PREDICTED: uncharacterized protein LOC105642006 isoform X3 [Jatropha curcas] |
| 13 |
Hb_000768_120 |
0.1439669507 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigB [Jatropha curcas] |
| 14 |
Hb_006681_020 |
0.1449809844 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP38, chloroplastic [Jatropha curcas] |
| 15 |
Hb_007479_010 |
0.145938367 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 16 |
Hb_004007_170 |
0.148511822 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 17 |
Hb_006583_030 |
0.1495507638 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like isoform X4 [Jatropha curcas] |
| 18 |
Hb_004855_070 |
0.1514314074 |
- |
- |
PLASTID-SPECIFIC RIBOSOMAL protein 4 [Populus trichocarpa] |
| 19 |
Hb_002027_100 |
0.1524482075 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000574_380 |
0.1542972591 |
- |
- |
hydrolase, putative [Ricinus communis] |