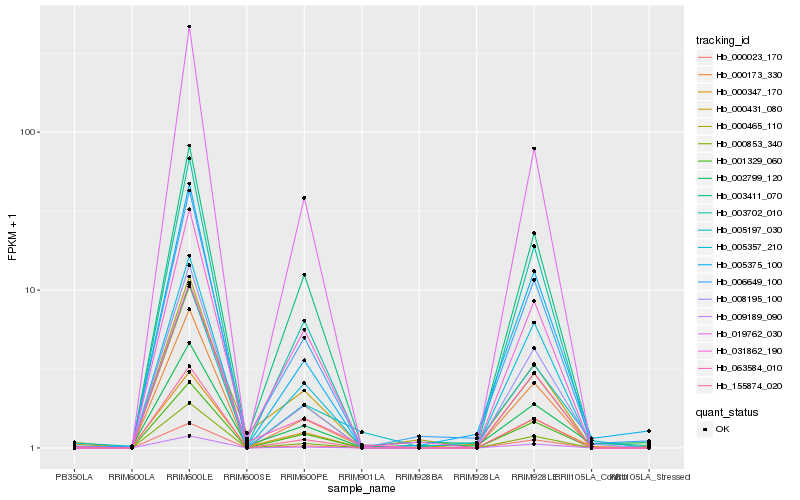
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005197_030 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH71-like isoform X2 [Populus euphratica] |
| 2 |
Hb_006649_100 |
0.131118186 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 3 |
Hb_000173_330 |
0.1332906643 |
- |
- |
PREDICTED: glutamine synthetase leaf isozyme, chloroplastic [Vitis vinifera] |
| 4 |
Hb_005375_100 |
0.1366407881 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH57-like [Jatropha curcas] |
| 5 |
Hb_000347_170 |
0.1367859496 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 6 |
Hb_008195_100 |
0.1384040136 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: protein YABBY 4 [Vitis vinifera] |
| 7 |
Hb_063584_010 |
0.1435566074 |
- |
- |
hypothetical protein JCGZ_10530 [Jatropha curcas] |
| 8 |
Hb_003411_070 |
0.1440934408 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 3.1 [Populus euphratica] |
| 9 |
Hb_003702_010 |
0.1481827436 |
- |
- |
PREDICTED: phospholipase A1-II 1-like [Jatropha curcas] |
| 10 |
Hb_000853_340 |
0.149994324 |
- |
- |
PREDICTED: kynurenine--oxoglutarate transaminase 1-like isoform X2 [Populus euphratica] |
| 11 |
Hb_000465_110 |
0.1501015569 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 330-like [Jatropha curcas] |
| 12 |
Hb_002799_120 |
0.1503976146 |
- |
- |
PREDICTED: uncharacterized protein LOC105141197 [Populus euphratica] |
| 13 |
Hb_000023_170 |
0.1514501231 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2-like [Jatropha curcas] |
| 14 |
Hb_155874_020 |
0.1525486984 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 15 |
Hb_005357_210 |
0.1534058875 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein MERISTEM L1 [Jatropha curcas] |
| 16 |
Hb_031862_190 |
0.1551866995 |
- |
- |
- |
| 17 |
Hb_009189_090 |
0.1553799235 |
- |
- |
protein phosphatase-2c, putative [Ricinus communis] |
| 18 |
Hb_001329_060 |
0.1558590371 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 19 |
Hb_000431_080 |
0.1559924951 |
- |
- |
PREDICTED: uncharacterized protein LOC105632547 [Jatropha curcas] |
| 20 |
Hb_019762_030 |
0.15644675 |
- |
- |
hypothetical protein POPTR_0002s08720g [Populus trichocarpa] |