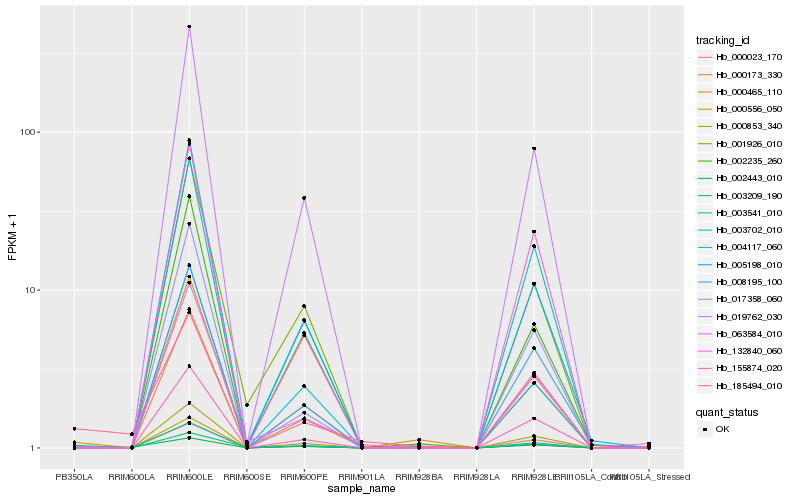
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003541_010 |
0.0 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_000465_110 |
0.1626139142 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 330-like [Jatropha curcas] |
| 3 |
Hb_185494_010 |
0.1808173031 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 4 |
Hb_003209_190 |
0.1839798375 |
- |
- |
hypothetical protein POPTR_0010s16770g [Populus trichocarpa] |
| 5 |
Hb_019762_030 |
0.1855618567 |
- |
- |
hypothetical protein POPTR_0002s08720g [Populus trichocarpa] |
| 6 |
Hb_000853_340 |
0.1862093665 |
- |
- |
PREDICTED: kynurenine--oxoglutarate transaminase 1-like isoform X2 [Populus euphratica] |
| 7 |
Hb_063584_010 |
0.1909869262 |
- |
- |
hypothetical protein JCGZ_10530 [Jatropha curcas] |
| 8 |
Hb_155874_020 |
0.1924259954 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 9 |
Hb_002443_010 |
0.1934180013 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 10 |
Hb_003702_010 |
0.1962964042 |
- |
- |
PREDICTED: phospholipase A1-II 1-like [Jatropha curcas] |
| 11 |
Hb_005198_010 |
0.1964409584 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 5 [Jatropha curcas] |
| 12 |
Hb_008195_100 |
0.1967013043 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: protein YABBY 4 [Vitis vinifera] |
| 13 |
Hb_002235_260 |
0.1981244798 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0017s12230g [Populus trichocarpa] |
| 14 |
Hb_000173_330 |
0.1982503043 |
- |
- |
PREDICTED: glutamine synthetase leaf isozyme, chloroplastic [Vitis vinifera] |
| 15 |
Hb_004117_060 |
0.198880661 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g33811 [Jatropha curcas] |
| 16 |
Hb_000023_170 |
0.2009246986 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2-like [Jatropha curcas] |
| 17 |
Hb_000556_050 |
0.201326352 |
- |
- |
- |
| 18 |
Hb_132840_060 |
0.2013507865 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH57-like [Jatropha curcas] |
| 19 |
Hb_017358_060 |
0.2023451949 |
- |
- |
PREDICTED: uncharacterized protein LOC105641510 [Jatropha curcas] |
| 20 |
Hb_001926_010 |
0.2028129737 |
- |
- |
hypothetical protein Csa_2G383910 [Cucumis sativus] |