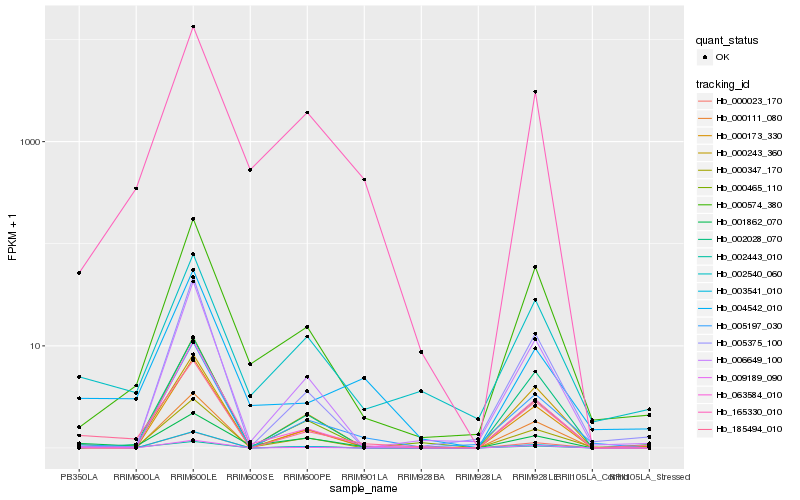
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_185494_010 |
0.0 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 2 |
Hb_002443_010 |
0.1515843413 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 3 |
Hb_002540_060 |
0.1617801245 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000574_380 |
0.1726554461 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 5 |
Hb_005197_030 |
0.1760558559 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH71-like isoform X2 [Populus euphratica] |
| 6 |
Hb_003541_010 |
0.1808173031 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_000347_170 |
0.1863233639 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 8 |
Hb_000173_330 |
0.1880737548 |
- |
- |
PREDICTED: glutamine synthetase leaf isozyme, chloroplastic [Vitis vinifera] |
| 9 |
Hb_004542_010 |
0.1943264924 |
- |
- |
Putative chloroplast RF19 [Medicago truncatula] |
| 10 |
Hb_000465_110 |
0.1956745088 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 330-like [Jatropha curcas] |
| 11 |
Hb_002028_070 |
0.1959479437 |
- |
- |
PREDICTED: circadian locomoter output cycles protein kaput [Jatropha curcas] |
| 12 |
Hb_005375_100 |
0.1964412178 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH57-like [Jatropha curcas] |
| 13 |
Hb_000111_080 |
0.1971547424 |
- |
- |
hypothetical protein JCGZ_24534 [Jatropha curcas] |
| 14 |
Hb_006649_100 |
0.198336825 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 15 |
Hb_165330_010 |
0.1991065881 |
- |
- |
BnaUnng00840D [Brassica napus] |
| 16 |
Hb_000243_360 |
0.2008389218 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 isoform X1 [Vitis vinifera] |
| 17 |
Hb_001862_070 |
0.2021932603 |
- |
- |
hypothetical protein RCOM_1155060 [Ricinus communis] |
| 18 |
Hb_000023_170 |
0.2032801407 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2-like [Jatropha curcas] |
| 19 |
Hb_009189_090 |
0.2034567388 |
- |
- |
protein phosphatase-2c, putative [Ricinus communis] |
| 20 |
Hb_063584_010 |
0.2040406476 |
- |
- |
hypothetical protein JCGZ_10530 [Jatropha curcas] |