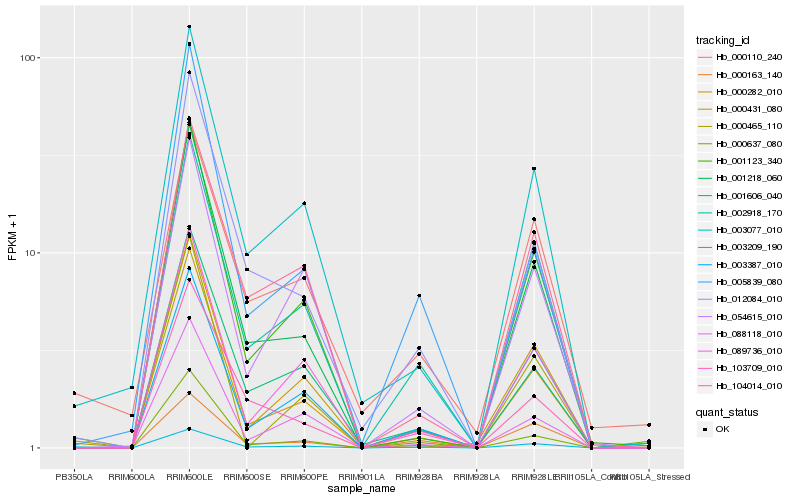
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003209_190 |
0.0 |
- |
- |
hypothetical protein POPTR_0010s16770g [Populus trichocarpa] |
| 2 |
Hb_002918_170 |
0.1430686018 |
- |
- |
PREDICTED: MACPF domain-containing protein NSL1 [Jatropha curcas] |
| 3 |
Hb_003077_010 |
0.1517728212 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit O, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_003387_010 |
0.1562275519 |
- |
- |
PREDICTED: protein kinase PINOID 2 [Gossypium raimondii] |
| 5 |
Hb_000110_240 |
0.1594673954 |
- |
- |
PREDICTED: uncharacterized protein LOC105642006 isoform X3 [Jatropha curcas] |
| 6 |
Hb_012084_010 |
0.162479748 |
- |
- |
cold regulated protein [Manihot esculenta] |
| 7 |
Hb_001218_060 |
0.1653525849 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_089736_010 |
0.1656719897 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 7 [Populus euphratica] |
| 9 |
Hb_001123_340 |
0.1723872722 |
- |
- |
PREDICTED: uncharacterized protein LOC105123089 isoform X1 [Populus euphratica] |
| 10 |
Hb_000465_110 |
0.1731630959 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 330-like [Jatropha curcas] |
| 11 |
Hb_000431_080 |
0.1765812188 |
- |
- |
PREDICTED: uncharacterized protein LOC105632547 [Jatropha curcas] |
| 12 |
Hb_001606_040 |
0.1785146536 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 13 |
Hb_000637_080 |
0.1801782279 |
- |
- |
PREDICTED: protein YLS3 [Jatropha curcas] |
| 14 |
Hb_000163_140 |
0.1804134761 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 15 |
Hb_000282_010 |
0.1804897925 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_054615_010 |
0.1809069274 |
- |
- |
Chromosome-associated kinesin KIF4A, putative [Ricinus communis] |
| 17 |
Hb_005839_080 |
0.1810313962 |
- |
- |
Transcription factor bHLH61 isoform 1 [Theobroma cacao] |
| 18 |
Hb_088118_010 |
0.1818177115 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 19 |
Hb_104014_010 |
0.1825989081 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39020 isoform X2 [Jatropha curcas] |
| 20 |
Hb_103709_010 |
0.183092251 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |