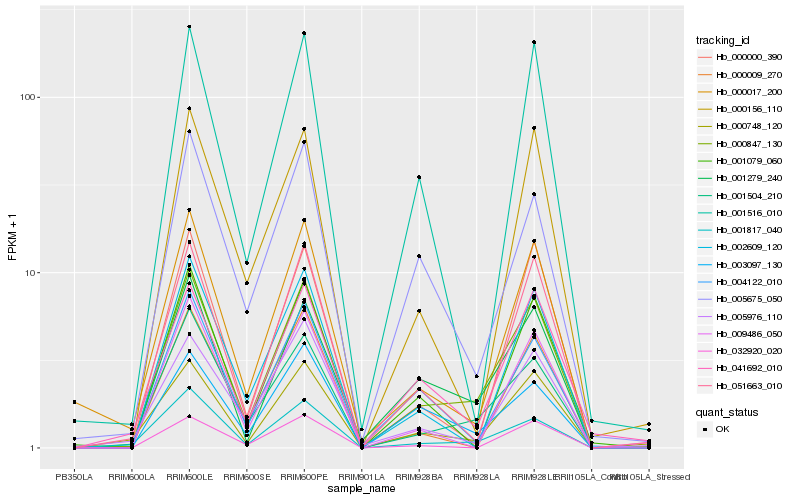
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000847_130 |
0.0 |
- |
- |
PREDICTED: fimbrin-2 [Jatropha curcas] |
| 2 |
Hb_001504_210 |
0.0982194897 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001279_240 |
0.1133725809 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105633500 [Jatropha curcas] |
| 4 |
Hb_001817_040 |
0.1205085865 |
- |
- |
myosin XI, putative [Ricinus communis] |
| 5 |
Hb_051663_010 |
0.12389986 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 6 |
Hb_000748_120 |
0.1244680144 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL1 [Jatropha curcas] |
| 7 |
Hb_000017_200 |
0.125288484 |
- |
- |
PREDICTED: uncharacterized protein LOC105645766 [Jatropha curcas] |
| 8 |
Hb_001516_010 |
0.1362550572 |
- |
- |
PREDICTED: guanylate kinase 2 [Jatropha curcas] |
| 9 |
Hb_001079_060 |
0.1388050481 |
- |
- |
PREDICTED: probable vacuolar amino acid transporter YPQ1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002609_120 |
0.1393245676 |
- |
- |
hypothetical protein POPTR_0001s25660g [Populus trichocarpa] |
| 11 |
Hb_004122_010 |
0.1405251754 |
- |
- |
PREDICTED: uncharacterized protein LOC105630923 [Jatropha curcas] |
| 12 |
Hb_000000_390 |
0.1406758314 |
- |
- |
PREDICTED: uncharacterized protein LOC105633753 [Jatropha curcas] |
| 13 |
Hb_005675_050 |
0.1406945623 |
- |
- |
alpha-glucosidase, putative [Ricinus communis] |
| 14 |
Hb_003097_130 |
0.1409571721 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_041692_010 |
0.1427656617 |
- |
- |
annexin [Manihot esculenta] |
| 16 |
Hb_000009_270 |
0.1443251859 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 17 |
Hb_000156_110 |
0.1453705617 |
- |
- |
Uncharacterized protein TCM_029490 [Theobroma cacao] |
| 18 |
Hb_032920_020 |
0.1456359615 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 19 |
Hb_005976_110 |
0.1461373397 |
- |
- |
PREDICTED: uncharacterized protein LOC105639708 [Jatropha curcas] |
| 20 |
Hb_009486_050 |
0.1461808681 |
- |
- |
PREDICTED: cyclin-U4-1 [Jatropha curcas] |