| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
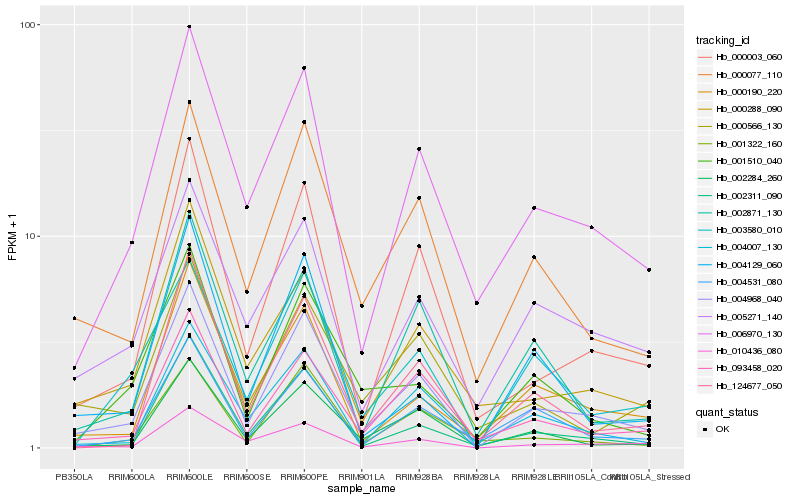
Hb_004968_040 |
0.0 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 5 [Jatropha curcas] |
| 2 |
Hb_000003_060 |
0.1145387391 |
transcription factor |
TF Family: HMG |
DNA-binding protein MNB1B, putative [Ricinus communis] |
| 3 |
Hb_000077_110 |
0.1218275983 |
- |
- |
PREDICTED: uncharacterized protein LOC105646935 [Jatropha curcas] |
| 4 |
Hb_093458_020 |
0.1234325453 |
desease resistance |
Gene Name: AAA |
PREDICTED: chromosome transmission fidelity protein 18 homolog [Jatropha curcas] |
| 5 |
Hb_002284_260 |
0.1297714363 |
- |
- |
brca1 interacting protein helicase 1 brip1, putative [Ricinus communis] |
| 6 |
Hb_006970_130 |
0.1299613566 |
- |
- |
PREDICTED: histone H3.3-like [Colobus angolensis palliatus] |
| 7 |
Hb_010436_080 |
0.1304151225 |
- |
- |
PREDICTED: DNA replication licensing factor MCM5 [Jatropha curcas] |
| 8 |
Hb_004007_130 |
0.1370115792 |
- |
- |
PREDICTED: uncharacterized protein LOC105639843 [Jatropha curcas] |
| 9 |
Hb_000190_220 |
0.1416335395 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek2 [Jatropha curcas] |
| 10 |
Hb_002871_130 |
0.1479295985 |
- |
- |
PREDICTED: non-structural maintenance of chromosomes element 4 homolog A isoform X2 [Jatropha curcas] |
| 11 |
Hb_003580_010 |
0.1495331133 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 12 |
Hb_004531_080 |
0.1514725684 |
- |
- |
hypothetical protein RCOM_1491960 [Ricinus communis] |
| 13 |
Hb_002311_090 |
0.151844609 |
- |
- |
PREDICTED: uncharacterized protein LOC105643503 [Jatropha curcas] |
| 14 |
Hb_000288_090 |
0.15574048 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_124677_050 |
0.1584229854 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000566_130 |
0.158689003 |
- |
- |
PREDICTED: probable DNA primase large subunit isoform X2 [Jatropha curcas] |
| 17 |
Hb_001510_040 |
0.1597463136 |
- |
- |
PREDICTED: 125 kDa kinesin-related protein [Jatropha curcas] |
| 18 |
Hb_004129_060 |
0.1605904396 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 19 |
Hb_005271_140 |
0.1609909916 |
transcription factor |
TF Family: LIM |
Pollen-specific protein SF3 [Morus notabilis] |
| 20 |
Hb_001322_160 |
0.1614993635 |
- |
- |
hypothetical protein JCGZ_03472 [Jatropha curcas] |