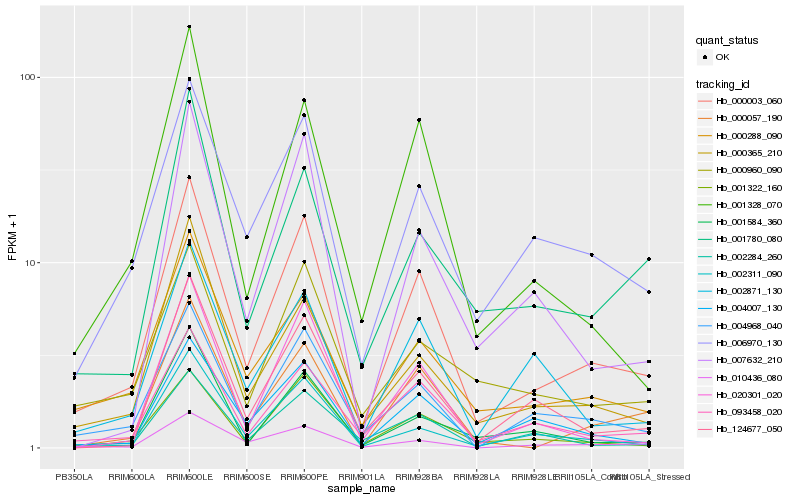
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_060 |
0.0 |
transcription factor |
TF Family: HMG |
DNA-binding protein MNB1B, putative [Ricinus communis] |
| 2 |
Hb_093458_020 |
0.1118900688 |
desease resistance |
Gene Name: AAA |
PREDICTED: chromosome transmission fidelity protein 18 homolog [Jatropha curcas] |
| 3 |
Hb_004968_040 |
0.1145387391 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 5 [Jatropha curcas] |
| 4 |
Hb_000288_090 |
0.1271700197 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_010436_080 |
0.128253284 |
- |
- |
PREDICTED: DNA replication licensing factor MCM5 [Jatropha curcas] |
| 6 |
Hb_006970_130 |
0.1303035077 |
- |
- |
PREDICTED: histone H3.3-like [Colobus angolensis palliatus] |
| 7 |
Hb_000057_190 |
0.1319838296 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004007_130 |
0.1336821501 |
- |
- |
PREDICTED: uncharacterized protein LOC105639843 [Jatropha curcas] |
| 9 |
Hb_001322_160 |
0.1401883039 |
- |
- |
hypothetical protein JCGZ_03472 [Jatropha curcas] |
| 10 |
Hb_001328_070 |
0.1413234775 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 13 [Jatropha curcas] |
| 11 |
Hb_000365_210 |
0.1423625898 |
- |
- |
PREDICTED: cell division control protein 2 homolog C [Jatropha curcas] |
| 12 |
Hb_002284_260 |
0.1451314874 |
- |
- |
brca1 interacting protein helicase 1 brip1, putative [Ricinus communis] |
| 13 |
Hb_002311_090 |
0.1465842558 |
- |
- |
PREDICTED: uncharacterized protein LOC105643503 [Jatropha curcas] |
| 14 |
Hb_020301_020 |
0.1499320698 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 15 |
Hb_124677_050 |
0.1537420605 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002871_130 |
0.155403328 |
- |
- |
PREDICTED: non-structural maintenance of chromosomes element 4 homolog A isoform X2 [Jatropha curcas] |
| 17 |
Hb_000960_090 |
0.1566281896 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_007632_210 |
0.1577141163 |
- |
- |
PREDICTED: sodium/potassium/calcium exchanger 1 [Populus euphratica] |
| 19 |
Hb_001780_080 |
0.1599607341 |
- |
- |
PREDICTED: tubulin beta-2 chain [Jatropha curcas] |
| 20 |
Hb_001584_360 |
0.1616664263 |
- |
- |
PREDICTED: serine/threonine-protein kinase haspin homolog hrk1 [Jatropha curcas] |